Gene
KWMTBOMO05479 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006709
Annotation
PREDICTED:_thioredoxin_domain-containing_protein_17-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.292
Sequence
CDS
ATGGTTATTAACGTGGATCTAAAAGGTTTTGAAGAATTTTCGAAATATACTCGTGCAATTGATTCTCGTGGTCCCCCTGTATTTTTTTACTTTAGCGGTTCAAAACTACCTGATGGCAATAGCTGGTGCCCCGACTGCGTGGAAGCTGAACCAGTCGTCAGGCATTACCTTAGTGAGCTCGACAAAAGTATCATTTTTGTCTATGTCGATGTTGGAGATAGAGAATATTGGAAGGACAAAGAGTGCCCGTTCAGAACGGACAGTCGCTCTAAGTTGATGGTCATACCGACTTTAATAAAATGGAAGGGAGTACAGAGGCTTGAGGGAAGCCAGTGCAGCAATCGGGAACTTCTGCAGATGTTGTTCGAAGAAGAAGATTAA
Protein
MVINVDLKGFEEFSKYTRAIDSRGPPVFFYFSGSKLPDGNSWCPDCVEAEPVVRHYLSELDKSIIFVYVDVGDREYWKDKECPFRTDSRSKLMVIPTLIKWKGVQRLEGSQCSNRELLQMLFEEED
Summary
Uniprot
H9JB14
A0A3S2NS99
A0A212FEU2
S4PI80
A0A194QHM1
A0A194RSL2
+ More
A0A2A4JB74 A0A0K0WTM1 A0A1E1WI61 A0A2H1VFA7 U5EK99 A0A1B0FPP7 A0A0L0CMC8 A0A0K8TNN5 V5GMV8 A0A1A9WKI2 A0A1A9VCC8 A0A1B0C1L8 A0A1A9Z7Z2 A0A1I8M158 A0A1A9XER9 A0A1Y1M931 A0A1A9X4U6 T1PFD3 D3TRX8 Q29NV0 B4P062 A0A1W4UHY6 A0A1Y1M1X6 A0A1Y1M6A0 A0A0C9QTK1 B4I1G5 B4Q3M1 A0A0J9QX63 B3N4W5 A0A0A1WWC4 A0A0Q5WGH9 A0A1L8DUB6 W8C3U6 A0A3B0K2R2 Q9VMQ9 A0A0M4ES68 B4MVJ2 B4KF30 B4LU19 A0A1I8PYG0 E9HMQ5 A0A0M4E385 A0A0N7ZDK8 A0A034VEI5 A0A0C9S0U9 A0A2R2YWV6 A0A182SLD6 B3MMY8 A0A1L8EIJ0 A0A1B0CZ90 A0A0K8VIG0 B4JAF9 A0A1W4WB29 A0A182K0G8 A0A182LYS8 A0A182R3S8 A0A182Q9I7 A0A1D6TL39 Q7Q1K7 A0A1D6TE11 A0A182P4Q6 A0A182XN80 A0A1S4H2T2 A0A182U4B1 A0A182V7S4 A0A182KU46 A0A182HRY0 A0A2M4C3G1 A0A182NBQ6 A0A2M3ZIM4 A0A182Y0M8 A0A182WGI2 A0A084WU38 A0A2M4C313 A0A182IVW6 W5JIF9 A0A182IZQ1 A0A2M4ARS7 A0A182F0Y0 A0A386R1D6 A0A0L7R8T5 A0A1L8DDS1 A0A1B0DL01 A0A1L8DCV6 A0A1L8DCV9 T1E849 A0A0A1WVH2 A0A139W9N0 A0A0M8ZPQ4 Q1HQU6 A0A2J7PD52 A0A154PNB0 A0A182H535 A0A1Q3FYI8
A0A2A4JB74 A0A0K0WTM1 A0A1E1WI61 A0A2H1VFA7 U5EK99 A0A1B0FPP7 A0A0L0CMC8 A0A0K8TNN5 V5GMV8 A0A1A9WKI2 A0A1A9VCC8 A0A1B0C1L8 A0A1A9Z7Z2 A0A1I8M158 A0A1A9XER9 A0A1Y1M931 A0A1A9X4U6 T1PFD3 D3TRX8 Q29NV0 B4P062 A0A1W4UHY6 A0A1Y1M1X6 A0A1Y1M6A0 A0A0C9QTK1 B4I1G5 B4Q3M1 A0A0J9QX63 B3N4W5 A0A0A1WWC4 A0A0Q5WGH9 A0A1L8DUB6 W8C3U6 A0A3B0K2R2 Q9VMQ9 A0A0M4ES68 B4MVJ2 B4KF30 B4LU19 A0A1I8PYG0 E9HMQ5 A0A0M4E385 A0A0N7ZDK8 A0A034VEI5 A0A0C9S0U9 A0A2R2YWV6 A0A182SLD6 B3MMY8 A0A1L8EIJ0 A0A1B0CZ90 A0A0K8VIG0 B4JAF9 A0A1W4WB29 A0A182K0G8 A0A182LYS8 A0A182R3S8 A0A182Q9I7 A0A1D6TL39 Q7Q1K7 A0A1D6TE11 A0A182P4Q6 A0A182XN80 A0A1S4H2T2 A0A182U4B1 A0A182V7S4 A0A182KU46 A0A182HRY0 A0A2M4C3G1 A0A182NBQ6 A0A2M3ZIM4 A0A182Y0M8 A0A182WGI2 A0A084WU38 A0A2M4C313 A0A182IVW6 W5JIF9 A0A182IZQ1 A0A2M4ARS7 A0A182F0Y0 A0A386R1D6 A0A0L7R8T5 A0A1L8DDS1 A0A1B0DL01 A0A1L8DCV6 A0A1L8DCV9 T1E849 A0A0A1WVH2 A0A139W9N0 A0A0M8ZPQ4 Q1HQU6 A0A2J7PD52 A0A154PNB0 A0A182H535 A0A1Q3FYI8
Pubmed
19121390
22118469
23622113
26354079
26108605
26369729
+ More
25315136 28004739 20353571 15632085 17994087 17550304 22936249 25830018 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 25348373 12364791 20966253 25244985 24438588 20920257 23761445 18362917 19820115 17204158 17510324 26483478
25315136 28004739 20353571 15632085 17994087 17550304 22936249 25830018 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 25348373 12364791 20966253 25244985 24438588 20920257 23761445 18362917 19820115 17204158 17510324 26483478
EMBL
BABH01008443
RSAL01000267
RVE43237.1
AGBW02008896
OWR52275.1
GAIX01005500
+ More
JAA87060.1 KQ458793 KPJ05043.1 KQ459984 KPJ19056.1 NWSH01002033 PCG69357.1 KP771808 AKS26495.1 GDQN01004351 GDQN01003183 GDQN01001704 JAT86703.1 JAT87871.1 JAT89350.1 ODYU01002242 SOQ39461.1 GANO01001890 JAB57981.1 CCAG010008058 JRES01000186 KNC33503.1 GDAI01001601 JAI16002.1 GALX01005554 JAB62912.1 JXJN01024136 GEZM01042467 JAV79837.1 KA647482 AFP62111.1 EZ424180 ADD20456.1 CH379058 EAL34544.1 CM000157 EDW88927.2 GEZM01042474 JAV79834.1 GEZM01042475 GEZM01042472 JAV79835.1 GBYB01007044 JAG76811.1 CH480820 EDW54372.1 CM000361 CM002910 EDX03825.1 KMY88303.1 KMY88304.1 CH954177 EDV57867.1 GBXI01011351 JAD02941.1 KQS70181.1 GFDF01004074 JAV10010.1 GAMC01005899 JAC00657.1 OUUW01000006 SPP82180.1 AE014134 AY060849 AY113391 AF503572 KX531732 AAF52253.1 AAL28397.1 AAM29396.1 AAN03825.1 AGB92634.1 AGB92635.1 ANY27542.1 CP012523 ALC40125.1 CH963857 EDW75712.1 CH933807 EDW13013.2 CH940649 EDW65072.1 GL732689 EFX66964.1 ALC38034.1 GDRN01031019 JAI67528.1 GAKP01018430 JAC40522.1 GBYB01014236 JAG84003.1 KR006967 AMK05619.1 CH902620 EDV31013.2 GFDG01000370 JAV18429.1 AJVK01002143 GDHF01013630 JAI38684.1 CH916368 EDW03830.1 AXCM01001134 AXCN02000684 AAAB01008980 EAA14229.2 APCN01001775 GGFJ01010570 MBW59711.1 GGFM01007643 MBW28394.1 ATLV01026995 KE525421 KFB53732.1 GGFJ01010569 MBW59710.1 ADMH02001427 ETN62579.1 GGFK01009977 MBW43298.1 MH143602 QCYY01001515 AYE56135.1 ROT77504.1 KQ414632 KOC67181.1 GFDF01009604 JAV04480.1 AJVK01068838 GFDF01009791 JAV04293.1 GFDF01009792 JAV04292.1 GAMD01003073 JAA98517.1 GBXI01011884 JAD02408.1 KQ972166 KYB24618.1 KQ435922 KOX68565.1 DQ440348 CH477855 ABF18381.1 EAT35597.1 NEVH01026394 PNF14252.1 KQ434998 KZC13322.1 JXUM01110751 KQ565441 KXJ70971.1 GFDL01002503 JAV32542.1
JAA87060.1 KQ458793 KPJ05043.1 KQ459984 KPJ19056.1 NWSH01002033 PCG69357.1 KP771808 AKS26495.1 GDQN01004351 GDQN01003183 GDQN01001704 JAT86703.1 JAT87871.1 JAT89350.1 ODYU01002242 SOQ39461.1 GANO01001890 JAB57981.1 CCAG010008058 JRES01000186 KNC33503.1 GDAI01001601 JAI16002.1 GALX01005554 JAB62912.1 JXJN01024136 GEZM01042467 JAV79837.1 KA647482 AFP62111.1 EZ424180 ADD20456.1 CH379058 EAL34544.1 CM000157 EDW88927.2 GEZM01042474 JAV79834.1 GEZM01042475 GEZM01042472 JAV79835.1 GBYB01007044 JAG76811.1 CH480820 EDW54372.1 CM000361 CM002910 EDX03825.1 KMY88303.1 KMY88304.1 CH954177 EDV57867.1 GBXI01011351 JAD02941.1 KQS70181.1 GFDF01004074 JAV10010.1 GAMC01005899 JAC00657.1 OUUW01000006 SPP82180.1 AE014134 AY060849 AY113391 AF503572 KX531732 AAF52253.1 AAL28397.1 AAM29396.1 AAN03825.1 AGB92634.1 AGB92635.1 ANY27542.1 CP012523 ALC40125.1 CH963857 EDW75712.1 CH933807 EDW13013.2 CH940649 EDW65072.1 GL732689 EFX66964.1 ALC38034.1 GDRN01031019 JAI67528.1 GAKP01018430 JAC40522.1 GBYB01014236 JAG84003.1 KR006967 AMK05619.1 CH902620 EDV31013.2 GFDG01000370 JAV18429.1 AJVK01002143 GDHF01013630 JAI38684.1 CH916368 EDW03830.1 AXCM01001134 AXCN02000684 AAAB01008980 EAA14229.2 APCN01001775 GGFJ01010570 MBW59711.1 GGFM01007643 MBW28394.1 ATLV01026995 KE525421 KFB53732.1 GGFJ01010569 MBW59710.1 ADMH02001427 ETN62579.1 GGFK01009977 MBW43298.1 MH143602 QCYY01001515 AYE56135.1 ROT77504.1 KQ414632 KOC67181.1 GFDF01009604 JAV04480.1 AJVK01068838 GFDF01009791 JAV04293.1 GFDF01009792 JAV04292.1 GAMD01003073 JAA98517.1 GBXI01011884 JAD02408.1 KQ972166 KYB24618.1 KQ435922 KOX68565.1 DQ440348 CH477855 ABF18381.1 EAT35597.1 NEVH01026394 PNF14252.1 KQ434998 KZC13322.1 JXUM01110751 KQ565441 KXJ70971.1 GFDL01002503 JAV32542.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000218220
+ More
UP000092444 UP000037069 UP000091820 UP000078200 UP000092460 UP000092445 UP000095301 UP000092443 UP000001819 UP000002282 UP000192221 UP000001292 UP000000304 UP000008711 UP000268350 UP000000803 UP000092553 UP000007798 UP000009192 UP000008792 UP000095300 UP000000305 UP000075901 UP000007801 UP000092462 UP000001070 UP000192223 UP000075881 UP000075883 UP000075900 UP000075886 UP000076407 UP000007062 UP000075902 UP000075885 UP000075903 UP000075882 UP000075840 UP000075884 UP000076408 UP000075920 UP000030765 UP000075880 UP000000673 UP000069272 UP000283509 UP000053825 UP000007266 UP000053105 UP000008820 UP000235965 UP000076502 UP000069940 UP000249989
UP000092444 UP000037069 UP000091820 UP000078200 UP000092460 UP000092445 UP000095301 UP000092443 UP000001819 UP000002282 UP000192221 UP000001292 UP000000304 UP000008711 UP000268350 UP000000803 UP000092553 UP000007798 UP000009192 UP000008792 UP000095300 UP000000305 UP000075901 UP000007801 UP000092462 UP000001070 UP000192223 UP000075881 UP000075883 UP000075900 UP000075886 UP000076407 UP000007062 UP000075902 UP000075885 UP000075903 UP000075882 UP000075840 UP000075884 UP000076408 UP000075920 UP000030765 UP000075880 UP000000673 UP000069272 UP000283509 UP000053825 UP000007266 UP000053105 UP000008820 UP000235965 UP000076502 UP000069940 UP000249989
Pfam
PF06110 DUF953
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
H9JB14
A0A3S2NS99
A0A212FEU2
S4PI80
A0A194QHM1
A0A194RSL2
+ More
A0A2A4JB74 A0A0K0WTM1 A0A1E1WI61 A0A2H1VFA7 U5EK99 A0A1B0FPP7 A0A0L0CMC8 A0A0K8TNN5 V5GMV8 A0A1A9WKI2 A0A1A9VCC8 A0A1B0C1L8 A0A1A9Z7Z2 A0A1I8M158 A0A1A9XER9 A0A1Y1M931 A0A1A9X4U6 T1PFD3 D3TRX8 Q29NV0 B4P062 A0A1W4UHY6 A0A1Y1M1X6 A0A1Y1M6A0 A0A0C9QTK1 B4I1G5 B4Q3M1 A0A0J9QX63 B3N4W5 A0A0A1WWC4 A0A0Q5WGH9 A0A1L8DUB6 W8C3U6 A0A3B0K2R2 Q9VMQ9 A0A0M4ES68 B4MVJ2 B4KF30 B4LU19 A0A1I8PYG0 E9HMQ5 A0A0M4E385 A0A0N7ZDK8 A0A034VEI5 A0A0C9S0U9 A0A2R2YWV6 A0A182SLD6 B3MMY8 A0A1L8EIJ0 A0A1B0CZ90 A0A0K8VIG0 B4JAF9 A0A1W4WB29 A0A182K0G8 A0A182LYS8 A0A182R3S8 A0A182Q9I7 A0A1D6TL39 Q7Q1K7 A0A1D6TE11 A0A182P4Q6 A0A182XN80 A0A1S4H2T2 A0A182U4B1 A0A182V7S4 A0A182KU46 A0A182HRY0 A0A2M4C3G1 A0A182NBQ6 A0A2M3ZIM4 A0A182Y0M8 A0A182WGI2 A0A084WU38 A0A2M4C313 A0A182IVW6 W5JIF9 A0A182IZQ1 A0A2M4ARS7 A0A182F0Y0 A0A386R1D6 A0A0L7R8T5 A0A1L8DDS1 A0A1B0DL01 A0A1L8DCV6 A0A1L8DCV9 T1E849 A0A0A1WVH2 A0A139W9N0 A0A0M8ZPQ4 Q1HQU6 A0A2J7PD52 A0A154PNB0 A0A182H535 A0A1Q3FYI8
A0A2A4JB74 A0A0K0WTM1 A0A1E1WI61 A0A2H1VFA7 U5EK99 A0A1B0FPP7 A0A0L0CMC8 A0A0K8TNN5 V5GMV8 A0A1A9WKI2 A0A1A9VCC8 A0A1B0C1L8 A0A1A9Z7Z2 A0A1I8M158 A0A1A9XER9 A0A1Y1M931 A0A1A9X4U6 T1PFD3 D3TRX8 Q29NV0 B4P062 A0A1W4UHY6 A0A1Y1M1X6 A0A1Y1M6A0 A0A0C9QTK1 B4I1G5 B4Q3M1 A0A0J9QX63 B3N4W5 A0A0A1WWC4 A0A0Q5WGH9 A0A1L8DUB6 W8C3U6 A0A3B0K2R2 Q9VMQ9 A0A0M4ES68 B4MVJ2 B4KF30 B4LU19 A0A1I8PYG0 E9HMQ5 A0A0M4E385 A0A0N7ZDK8 A0A034VEI5 A0A0C9S0U9 A0A2R2YWV6 A0A182SLD6 B3MMY8 A0A1L8EIJ0 A0A1B0CZ90 A0A0K8VIG0 B4JAF9 A0A1W4WB29 A0A182K0G8 A0A182LYS8 A0A182R3S8 A0A182Q9I7 A0A1D6TL39 Q7Q1K7 A0A1D6TE11 A0A182P4Q6 A0A182XN80 A0A1S4H2T2 A0A182U4B1 A0A182V7S4 A0A182KU46 A0A182HRY0 A0A2M4C3G1 A0A182NBQ6 A0A2M3ZIM4 A0A182Y0M8 A0A182WGI2 A0A084WU38 A0A2M4C313 A0A182IVW6 W5JIF9 A0A182IZQ1 A0A2M4ARS7 A0A182F0Y0 A0A386R1D6 A0A0L7R8T5 A0A1L8DDS1 A0A1B0DL01 A0A1L8DCV6 A0A1L8DCV9 T1E849 A0A0A1WVH2 A0A139W9N0 A0A0M8ZPQ4 Q1HQU6 A0A2J7PD52 A0A154PNB0 A0A182H535 A0A1Q3FYI8
PDB
1V9W
E-value=2.14114e-24,
Score=271
Ontologies
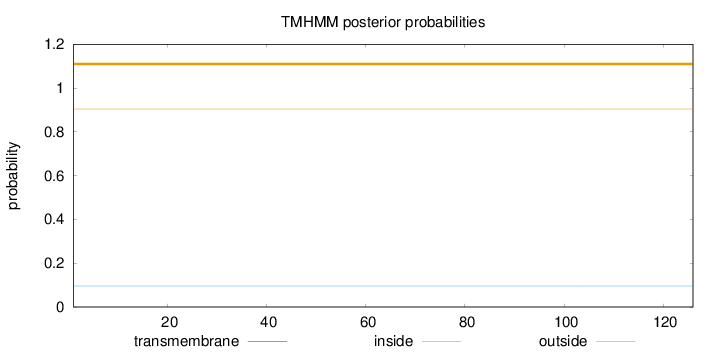
Topology
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09589
outside
1 - 126
Population Genetic Test Statistics
Pi
163.815622
Theta
149.58305
Tajima's D
0.338071
CLR
0
CSRT
0.467526623668817
Interpretation
Uncertain
