Gene
KWMTBOMO05472
Pre Gene Modal
BGIBMGA006633
Annotation
putative_axonemal_dynein_intermediate_chain_inner_arm_i1_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.627 Nuclear Reliability : 1.049
Sequence
CDS
ATGTCGTCCAATATAATGCTACCGGAATCCCCGGATCCCAATACACGATCAGAAACTGTAGCCTCAAAGATCAGTTTCAATGAGCAGTCTTCATCTAGAACAAGCAAAGTTGCGTTTTCGTTTATGGAGAAAAGAGCAATGTATCGTGTTGTGGTTGACGGTGCCGATGTCACACCCGAAGACATTCTCGAGCCTGATTATGTAATGATGGTAGATACTTTTTCGCACGCTGCCTTTGAAAACAAACCCCGTTTTAAAAGTGAAACCGGGCTTAAAGCTACGTCTAAATCACGATCCGTTACGGAGGGTGCGATGACTGTTTTTGCAACGACAATCTCTCTTGACGACATTCATATCGATCACACGCTCAACACGAGCGAGGGTTTTATACCTGACGACGAAGTTCTCGAACAAGCGCCTTCAGCATTTTATCTCCCGAAAAGTAAACCAGTCATGTCTTATCCCCCAGAAATTTTCATTGTGCTTAAAGAGACAGAAACTTGCTATTTGTTCGAATTACCTAGTTTTGCTTTTGAGAAAGGTACTCCTGAAGCAGATAAGGTAGAAGAGGATAATGAGTTTTATCAATATATTACTGTTGGAAAAGGGAGAAACAGAAAAATGATTGTGGAGGAAACACAAACCGAAGAGTATATTGTACAAACACGACACACGCTCGCGTCGAGACCCCAAAGGAAAAATGCTATAGCATTCGCATCTTTATGGGACATGCACGATACCTACGCTAAGCTTGCAAACGTGAAAAAAGTAGAAGAACCTGATGAAATGGTGCTCTATCAATCAGCAGCACCCAATTTACTCAGAAAGAAGAAACTAAATGTTTTTGACGCTTCCTTGAAAAAAGGAAAATGTTTTGAAGAACTAGCGTCGACACCAGAATTCCTAGATGCGGTTTTATTGGCTGAACGTGTATTATCGACACTAGAATATTCTGAAGCTCAGAAGAAATACCGTGGCTTGGTTCGTATAGATCCACTCTCGTTAGATTTAGTGTATATTTACACAATGAAACCTTTATGGACTTTTGAATGTGAAGATACCATAAACCGCCCCATAACTGGAATCTCATTTAATCCGAAGAATGAGAATATATTAGCAGTCGCTCACGGGAAGTTCCGGTATGCTGAATGCTATAACGGCTTGGTCTGTATTTGGTGTACAAAAAATCCACTAAAACCCGAAAGAGTTTACCAATTCGAAGATCCATTAACATCATTGGCATTTGCCGAAAAAAACCCTAACTGGCTAGCTTGCGGTTTTGCTAACGGTGACGTATTGATTTTGGACATCACTTCATACACCACAAAAAGCATAGCGAAAAGTAAACGCGACACAAATCCCTGTTTTGAACCAATCTGGGTGACAAGTTGGCGATCAATTGATAAGGATAATGAATTCGTTATGACTTCATGTCAAGACGGAAGGATCAACAAATTTACTAACACTAAGACCCACGATTTTATATGTTCACCCATGATGAGAATATCAACCGTCGAAGGTAAGATGAAAGGGCTGGAAGCAACGAAAGCATGTTTGAAAGTAGACGTTCCTATAACGCGCTACCCAGCAGCGTTATGCATAAAATGGCACCCGAACGTTGAGCACGTGTATCTAGTTGGTACTGATGAAGGATGTATTCACAAATGTTCTACTCATTATCTGAATCAACACATCGAGGTCTTCAAAGCACATGCAGGTCCTGTTTACGGCATGGAGTACTCCCCTTTTATGAGTTCGTTGTTGGTGTCTTGCGGCGCAGACGGTGCTATAAGATTGTGGATAGAAGGCATGGATGACGTCATTATGACCTTAACATGTCCCGCTGCAGTATACGGTATCGCATTCTGTCCAATCAACTCCACGGTGATTTTATCAGTAAGCGGAAATATACTTTCCATTTGGGATCTGCGAAGGAAAACTCATATGCCGTGTGCGGAATACTCTTTTCCTAATAACGTTGTTCTGACGTACATTAAATTTGCCGCTTCGGGAGATAATGTCTTCGTTGGTGAGACGTCTGGACGCATTCACACATTCCATTTGGAAGACACTCCGATACCACCGTTTTACCAAAGAAAGATGTTAGACGAGGCGATCAAAAAAGCACTTTGCACGAGACCACAGCTCTTGAAGCAACTCGAAAAATTGGAAAAATTTAGAGAGAAGTATGCGAAATAA
Protein
MSSNIMLPESPDPNTRSETVASKISFNEQSSSRTSKVAFSFMEKRAMYRVVVDGADVTPEDILEPDYVMMVDTFSHAAFENKPRFKSETGLKATSKSRSVTEGAMTVFATTISLDDIHIDHTLNTSEGFIPDDEVLEQAPSAFYLPKSKPVMSYPPEIFIVLKETETCYLFELPSFAFEKGTPEADKVEEDNEFYQYITVGKGRNRKMIVEETQTEEYIVQTRHTLASRPQRKNAIAFASLWDMHDTYAKLANVKKVEEPDEMVLYQSAAPNLLRKKKLNVFDASLKKGKCFEELASTPEFLDAVLLAERVLSTLEYSEAQKKYRGLVRIDPLSLDLVYIYTMKPLWTFECEDTINRPITGISFNPKNENILAVAHGKFRYAECYNGLVCIWCTKNPLKPERVYQFEDPLTSLAFAEKNPNWLACGFANGDVLILDITSYTTKSIAKSKRDTNPCFEPIWVTSWRSIDKDNEFVMTSCQDGRINKFTNTKTHDFICSPMMRISTVEGKMKGLEATKACLKVDVPITRYPAALCIKWHPNVEHVYLVGTDEGCIHKCSTHYLNQHIEVFKAHAGPVYGMEYSPFMSSLLVSCGADGAIRLWIEGMDDVIMTLTCPAAVYGIAFCPINSTVILSVSGNILSIWDLRRKTHMPCAEYSFPNNVVLTYIKFAASGDNVFVGETSGRIHTFHLEDTPIPPFYQRKMLDEAIKKALCTRPQLLKQLEKLEKFREKYAK
Summary
Similarity
Belongs to the universal ribosomal protein uL23 family.
Uniprot
H9JAT8
A0A0L7LF19
A0A1C9EGD9
A0A212FEX4
A0A194RML2
A0A194QNH2
+ More
A0A2A4JWC2 A0A2A4JXE5 A0A2H1W1J7 A0A2H1W1Z7 A0A2P8Z1A1 A0A195CCZ3 A0A151WSK9 E2AHR7 A0A0J7KJF0 A0A195FQ71 A0A310SYD6 A0A195BWE5 A0A026VSZ1 A0A158NUZ6 E9IDE1 U4UW31 A0A154PJ99 F4WXI0 A0A088AVI2 K7IPP0 A0A0L7RJ61 A0A232ENU0 A0A139WC78 A0A0N0U371 A0A1W4XIY1 Q175V4 A0A182GRE0 A0A182GBN8 A0A1Q3FSZ8 B0WT55 A0A1J1I5I4 A0A336MF52 A0A1B6EAA7 E0VLP7 A0A084W427 A0A1Y9IW76 A0A1I8P0R3 A0A182QUY5 A0A182FDI7 A0A182JJ59 A0A182ME34 A0A1A9WB47 A0A182YPL7 A0A182SAK2 A0A182PJU5 A0A2Y9D1H0 A0A1B6GFQ3 A0A1B6JS29 A0A1B0FBW5 A0A182VFI5 A0NEL3 A0A1A9UUL6 A0A182TX37 A0A182KPC1 A0A182K016 A0A0L0BME4 A0A182R737 A0A1B0AX94 A0A0L0BUB4 A0A182XM29 A0A182I1N9 A0A1A9WPR9 B4H3F4 Q2LZP2 A0A1I8N4X0 B4PDR9 B3NET5 B4HVY2 A0A0J9RM05 B4QMI8 A0A1I8MVP1 A0A1W4W687 D6W4W3 Q9W0A6 A0A3B0K0R9 B3M7R3 B4MND6 B4L966 Q29ET9 A0A1B0C359 A0A1A9V5V7 T1GR27 A0A1W4VNQ9 B4PC12 B4L903 A0A1I8PQ75 A0A0J9RKR0 B4HVB3
A0A2A4JWC2 A0A2A4JXE5 A0A2H1W1J7 A0A2H1W1Z7 A0A2P8Z1A1 A0A195CCZ3 A0A151WSK9 E2AHR7 A0A0J7KJF0 A0A195FQ71 A0A310SYD6 A0A195BWE5 A0A026VSZ1 A0A158NUZ6 E9IDE1 U4UW31 A0A154PJ99 F4WXI0 A0A088AVI2 K7IPP0 A0A0L7RJ61 A0A232ENU0 A0A139WC78 A0A0N0U371 A0A1W4XIY1 Q175V4 A0A182GRE0 A0A182GBN8 A0A1Q3FSZ8 B0WT55 A0A1J1I5I4 A0A336MF52 A0A1B6EAA7 E0VLP7 A0A084W427 A0A1Y9IW76 A0A1I8P0R3 A0A182QUY5 A0A182FDI7 A0A182JJ59 A0A182ME34 A0A1A9WB47 A0A182YPL7 A0A182SAK2 A0A182PJU5 A0A2Y9D1H0 A0A1B6GFQ3 A0A1B6JS29 A0A1B0FBW5 A0A182VFI5 A0NEL3 A0A1A9UUL6 A0A182TX37 A0A182KPC1 A0A182K016 A0A0L0BME4 A0A182R737 A0A1B0AX94 A0A0L0BUB4 A0A182XM29 A0A182I1N9 A0A1A9WPR9 B4H3F4 Q2LZP2 A0A1I8N4X0 B4PDR9 B3NET5 B4HVY2 A0A0J9RM05 B4QMI8 A0A1I8MVP1 A0A1W4W687 D6W4W3 Q9W0A6 A0A3B0K0R9 B3M7R3 B4MND6 B4L966 Q29ET9 A0A1B0C359 A0A1A9V5V7 T1GR27 A0A1W4VNQ9 B4PC12 B4L903 A0A1I8PQ75 A0A0J9RKR0 B4HVB3
Pubmed
19121390
26227816
22118469
26354079
29403074
20798317
+ More
24508170 30249741 21347285 21282665 23537049 21719571 20075255 28648823 18362917 19820115 17510324 26483478 20566863 24438588 25244985 12364791 14747013 17210077 20966253 26108605 17994087 15632085 23185243 25315136 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
24508170 30249741 21347285 21282665 23537049 21719571 20075255 28648823 18362917 19820115 17510324 26483478 20566863 24438588 25244985 12364791 14747013 17210077 20966253 26108605 17994087 15632085 23185243 25315136 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867
EMBL
BABH01008440
JTDY01001470
KOB73786.1
KX778609
AON96557.1
AGBW02008896
+ More
OWR52268.1 KQ459984 KPJ19063.1 KQ458793 KPJ05036.1 NWSH01000454 PCG76301.1 PCG76304.1 ODYU01005770 SOQ46975.1 SOQ46976.1 PYGN01000245 PSN50258.1 KQ978023 KYM97953.1 KQ982769 KYQ50889.1 GL439579 EFN66996.1 LBMM01006506 KMQ90553.1 KQ981335 KYN42623.1 KQ759817 OAD62764.1 KQ976401 KYM92605.1 KK108145 QOIP01000010 EZA46852.1 RLU17915.1 ADTU01026863 GL762454 EFZ21532.1 KB632390 ERL94430.1 KQ434936 KZC11883.1 GL888425 EGI61050.1 KQ414582 KOC70895.1 NNAY01003071 OXU20024.1 KQ971371 KYB25578.1 KQ435922 KOX68482.1 CH477395 EAT41851.1 JXUM01082292 JXUM01082293 KQ563302 KXJ74096.1 JXUM01009334 KQ560281 KXJ83326.1 GFDL01004314 JAV30731.1 DS232080 EDS34198.1 CVRI01000042 CRK95557.1 UFQT01001082 SSX28796.1 GEDC01002429 JAS34869.1 DS235278 EEB14303.1 ATLV01020271 KE525297 KFB44971.1 AXCN02000403 AXCM01001951 GECZ01008511 JAS61258.1 GECU01005691 JAT02016.1 CCAG010023863 AAAB01008960 EAU76534.2 JRES01001641 KNC21300.1 JXJN01005154 JRES01001325 KNC23611.1 APCN01000070 CH479206 EDW30894.1 CH379069 EAL29465.2 KRT08035.1 CM000159 EDW92884.1 CH954178 EDV50208.1 CH480817 EDW50097.1 CM002912 KMY96921.1 CM000363 EDX08845.1 BT125013 ADI46809.1 AE014296 AAF47547.2 OUUW01000002 SPP76958.1 CH902618 EDV38786.1 CH963847 EDW72645.1 CH933816 EDW17241.1 CH379070 EAL29971.2 JXJN01024847 CAQQ02080280 EDW92667.1 EDW17178.1 KMY96563.1 EDW49878.1
OWR52268.1 KQ459984 KPJ19063.1 KQ458793 KPJ05036.1 NWSH01000454 PCG76301.1 PCG76304.1 ODYU01005770 SOQ46975.1 SOQ46976.1 PYGN01000245 PSN50258.1 KQ978023 KYM97953.1 KQ982769 KYQ50889.1 GL439579 EFN66996.1 LBMM01006506 KMQ90553.1 KQ981335 KYN42623.1 KQ759817 OAD62764.1 KQ976401 KYM92605.1 KK108145 QOIP01000010 EZA46852.1 RLU17915.1 ADTU01026863 GL762454 EFZ21532.1 KB632390 ERL94430.1 KQ434936 KZC11883.1 GL888425 EGI61050.1 KQ414582 KOC70895.1 NNAY01003071 OXU20024.1 KQ971371 KYB25578.1 KQ435922 KOX68482.1 CH477395 EAT41851.1 JXUM01082292 JXUM01082293 KQ563302 KXJ74096.1 JXUM01009334 KQ560281 KXJ83326.1 GFDL01004314 JAV30731.1 DS232080 EDS34198.1 CVRI01000042 CRK95557.1 UFQT01001082 SSX28796.1 GEDC01002429 JAS34869.1 DS235278 EEB14303.1 ATLV01020271 KE525297 KFB44971.1 AXCN02000403 AXCM01001951 GECZ01008511 JAS61258.1 GECU01005691 JAT02016.1 CCAG010023863 AAAB01008960 EAU76534.2 JRES01001641 KNC21300.1 JXJN01005154 JRES01001325 KNC23611.1 APCN01000070 CH479206 EDW30894.1 CH379069 EAL29465.2 KRT08035.1 CM000159 EDW92884.1 CH954178 EDV50208.1 CH480817 EDW50097.1 CM002912 KMY96921.1 CM000363 EDX08845.1 BT125013 ADI46809.1 AE014296 AAF47547.2 OUUW01000002 SPP76958.1 CH902618 EDV38786.1 CH963847 EDW72645.1 CH933816 EDW17241.1 CH379070 EAL29971.2 JXJN01024847 CAQQ02080280 EDW92667.1 EDW17178.1 KMY96563.1 EDW49878.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000245037 UP000078542 UP000075809 UP000000311 UP000036403 UP000078541 UP000078540 UP000053097 UP000279307 UP000005205 UP000030742 UP000076502 UP000007755 UP000005203 UP000002358 UP000053825 UP000215335 UP000007266 UP000053105 UP000192223 UP000008820 UP000069940 UP000249989 UP000002320 UP000183832 UP000009046 UP000030765 UP000075920 UP000095300 UP000075886 UP000069272 UP000075880 UP000075883 UP000091820 UP000076408 UP000075901 UP000075885 UP000075884 UP000092444 UP000075903 UP000007062 UP000078200 UP000075902 UP000075882 UP000075881 UP000037069 UP000075900 UP000092460 UP000076407 UP000075840 UP000008744 UP000001819 UP000095301 UP000002282 UP000008711 UP000001292 UP000000304 UP000192221 UP000000803 UP000268350 UP000007801 UP000007798 UP000009192 UP000015102
UP000245037 UP000078542 UP000075809 UP000000311 UP000036403 UP000078541 UP000078540 UP000053097 UP000279307 UP000005205 UP000030742 UP000076502 UP000007755 UP000005203 UP000002358 UP000053825 UP000215335 UP000007266 UP000053105 UP000192223 UP000008820 UP000069940 UP000249989 UP000002320 UP000183832 UP000009046 UP000030765 UP000075920 UP000095300 UP000075886 UP000069272 UP000075880 UP000075883 UP000091820 UP000076408 UP000075901 UP000075885 UP000075884 UP000092444 UP000075903 UP000007062 UP000078200 UP000075902 UP000075882 UP000075881 UP000037069 UP000075900 UP000092460 UP000076407 UP000075840 UP000008744 UP000001819 UP000095301 UP000002282 UP000008711 UP000001292 UP000000304 UP000192221 UP000000803 UP000268350 UP000007801 UP000007798 UP000009192 UP000015102
Pfam
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR001806 Small_GTPase
IPR005225 Small_GTP-bd_dom
IPR001014 Ribosomal_L23/L25_CS
IPR012678 Ribosomal_L23/L15e_core_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR005633 Ribosomal_L23/L25_N
IPR013025 Ribosomal_L25/23
IPR001148 CA_dom
IPR036398 CA_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR019775 WD40_repeat_CS
IPR027417 P-loop_NTPase
IPR001806 Small_GTPase
IPR005225 Small_GTP-bd_dom
IPR001014 Ribosomal_L23/L25_CS
IPR012678 Ribosomal_L23/L15e_core_dom_sf
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR005633 Ribosomal_L23/L25_N
IPR013025 Ribosomal_L25/23
IPR001148 CA_dom
IPR036398 CA_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JAT8
A0A0L7LF19
A0A1C9EGD9
A0A212FEX4
A0A194RML2
A0A194QNH2
+ More
A0A2A4JWC2 A0A2A4JXE5 A0A2H1W1J7 A0A2H1W1Z7 A0A2P8Z1A1 A0A195CCZ3 A0A151WSK9 E2AHR7 A0A0J7KJF0 A0A195FQ71 A0A310SYD6 A0A195BWE5 A0A026VSZ1 A0A158NUZ6 E9IDE1 U4UW31 A0A154PJ99 F4WXI0 A0A088AVI2 K7IPP0 A0A0L7RJ61 A0A232ENU0 A0A139WC78 A0A0N0U371 A0A1W4XIY1 Q175V4 A0A182GRE0 A0A182GBN8 A0A1Q3FSZ8 B0WT55 A0A1J1I5I4 A0A336MF52 A0A1B6EAA7 E0VLP7 A0A084W427 A0A1Y9IW76 A0A1I8P0R3 A0A182QUY5 A0A182FDI7 A0A182JJ59 A0A182ME34 A0A1A9WB47 A0A182YPL7 A0A182SAK2 A0A182PJU5 A0A2Y9D1H0 A0A1B6GFQ3 A0A1B6JS29 A0A1B0FBW5 A0A182VFI5 A0NEL3 A0A1A9UUL6 A0A182TX37 A0A182KPC1 A0A182K016 A0A0L0BME4 A0A182R737 A0A1B0AX94 A0A0L0BUB4 A0A182XM29 A0A182I1N9 A0A1A9WPR9 B4H3F4 Q2LZP2 A0A1I8N4X0 B4PDR9 B3NET5 B4HVY2 A0A0J9RM05 B4QMI8 A0A1I8MVP1 A0A1W4W687 D6W4W3 Q9W0A6 A0A3B0K0R9 B3M7R3 B4MND6 B4L966 Q29ET9 A0A1B0C359 A0A1A9V5V7 T1GR27 A0A1W4VNQ9 B4PC12 B4L903 A0A1I8PQ75 A0A0J9RKR0 B4HVB3
A0A2A4JWC2 A0A2A4JXE5 A0A2H1W1J7 A0A2H1W1Z7 A0A2P8Z1A1 A0A195CCZ3 A0A151WSK9 E2AHR7 A0A0J7KJF0 A0A195FQ71 A0A310SYD6 A0A195BWE5 A0A026VSZ1 A0A158NUZ6 E9IDE1 U4UW31 A0A154PJ99 F4WXI0 A0A088AVI2 K7IPP0 A0A0L7RJ61 A0A232ENU0 A0A139WC78 A0A0N0U371 A0A1W4XIY1 Q175V4 A0A182GRE0 A0A182GBN8 A0A1Q3FSZ8 B0WT55 A0A1J1I5I4 A0A336MF52 A0A1B6EAA7 E0VLP7 A0A084W427 A0A1Y9IW76 A0A1I8P0R3 A0A182QUY5 A0A182FDI7 A0A182JJ59 A0A182ME34 A0A1A9WB47 A0A182YPL7 A0A182SAK2 A0A182PJU5 A0A2Y9D1H0 A0A1B6GFQ3 A0A1B6JS29 A0A1B0FBW5 A0A182VFI5 A0NEL3 A0A1A9UUL6 A0A182TX37 A0A182KPC1 A0A182K016 A0A0L0BME4 A0A182R737 A0A1B0AX94 A0A0L0BUB4 A0A182XM29 A0A182I1N9 A0A1A9WPR9 B4H3F4 Q2LZP2 A0A1I8N4X0 B4PDR9 B3NET5 B4HVY2 A0A0J9RM05 B4QMI8 A0A1I8MVP1 A0A1W4W687 D6W4W3 Q9W0A6 A0A3B0K0R9 B3M7R3 B4MND6 B4L966 Q29ET9 A0A1B0C359 A0A1A9V5V7 T1GR27 A0A1W4VNQ9 B4PC12 B4L903 A0A1I8PQ75 A0A0J9RKR0 B4HVB3
PDB
6F3A
E-value=3.46583e-12,
Score=176
Ontologies
GO
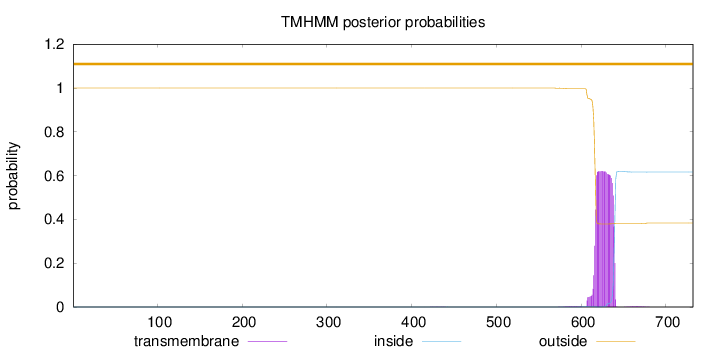
Topology
Length:
732
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.70625
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00025
outside
1 - 732
Population Genetic Test Statistics
Pi
325.325987
Theta
233.275602
Tajima's D
1.220923
CLR
0
CSRT
0.724763761811909
Interpretation
Possibly Positive selection
