Gene
KWMTBOMO05460
Pre Gene Modal
BGIBMGA006639
Annotation
PREDICTED:_glutathione_S-transferase_D7-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.158
Sequence
CDS
ATGTTACCTACGGTATTCGGAAGCCTCCAGGAGCCCACGGCGAAGCATCTAGAAGACATCGAGTCAGCGTACGGAGTAATCGAAGCGTACCTGCGGAAGCACCACTACATCGCGACCGACCACCTCACCATAGCCGACATCGCCGTTGGCTGCACCGCATTCGCTCTAAGAGGAGTGTTGGAAATTGATGAAAACAAGTTCCCAGGTATGATAAGATGGCTTCAAATTCTGAGCGAAGTTCCGAAAATCAAGAAACATGGTGTTTCTGGTGGTGATTTACTCGCAGATATATTAAGAAAACAGTGGAAAAAGAACAAGAATGTAGAAAAAATATATGATATTGATATATCCTATGGTATATCCTAG
Protein
MLPTVFGSLQEPTAKHLEDIESAYGVIEAYLRKHHYIATDHLTIADIAVGCTAFALRGVLEIDENKFPGMIRWLQILSEVPKIKKHGVSGGDLLADILRKQWKKNKNVEKIYDIDISYGIS
Summary
Similarity
Belongs to the GST superfamily.
Belongs to the glutamine synthetase family.
Belongs to the glutamine synthetase family.
Uniprot
H9JAU4
A0A3G1ZLC2
V5QP65
A0A2H1WDG8
A0A3G1ZLA8
D2I930
+ More
A0A0D3M5U3 A0A212FES8 A0A2H1WEV2 A0A1C9EGF0 A0A1C9EGF2 A0A075X2I2 A0A2A4IXK8 A0A0D3M5X3 A0A3G1T1G1 X5CCB3 A0A3G1ZLA9 A0A2H1WDA0 A0A2H1WD57 A0A0K8TUF8 D2I931 A0A3G1ZLB1 A0A0A7KL70 A0A2A4IV17 A0A194QNF8 A0A212F9V3 A0A194RSN3 A0A1W6I4K8 A0A212F3G0 A0A0K8TVL2 W8C6H4 Q7Z0Q7 Q7Z0Q9 T1PDX5 A0A1I8M6Z2 Q7Z0Q8 A9UIB0 A0A1W6I4K6 A0A034W9N4 A0A1L8EFG7 B4KJV1 A0A1L8EFD4 Q9U796 A0A0K8W5E9 A0A0D3M5V9 A0A0K8WMC3 H9JKA0 A0A0S3G4L2 A0A1I8PF33 B0LB15 A0A075X2I7 A0A1I8MHG2 T1PG87 A0A0A1WSG9 Q9U795 A0A0A1XQS3 A0A3G1ZLB6 A0A0J9RFT5 U5EZL1 A0A1L8EFQ5 A0A2A4JII4 K9LHE2 A0A2H1WRE6 A0A1I8Q311 A0A075X2V3 T1PFR3 A0A1W4UHU1 A0A3G1ZLC0 A0A3G1ZLA7 B4HNN4 A0A1L8D6F5 A0A2H1WIN4 A0A1I8MAH9 B4P4L9 A0A0L0CBD3 B4KTI4 A0A1P8PEU4 A0A1W4UG55 A0A1I8MXR0 A0A1I8M6H7 A0A125S6Q2 B4QCE3 A0A1A9WI52 B4QCE4 A0A182WH79 A0A034WM97
A0A0D3M5U3 A0A212FES8 A0A2H1WEV2 A0A1C9EGF0 A0A1C9EGF2 A0A075X2I2 A0A2A4IXK8 A0A0D3M5X3 A0A3G1T1G1 X5CCB3 A0A3G1ZLA9 A0A2H1WDA0 A0A2H1WD57 A0A0K8TUF8 D2I931 A0A3G1ZLB1 A0A0A7KL70 A0A2A4IV17 A0A194QNF8 A0A212F9V3 A0A194RSN3 A0A1W6I4K8 A0A212F3G0 A0A0K8TVL2 W8C6H4 Q7Z0Q7 Q7Z0Q9 T1PDX5 A0A1I8M6Z2 Q7Z0Q8 A9UIB0 A0A1W6I4K6 A0A034W9N4 A0A1L8EFG7 B4KJV1 A0A1L8EFD4 Q9U796 A0A0K8W5E9 A0A0D3M5V9 A0A0K8WMC3 H9JKA0 A0A0S3G4L2 A0A1I8PF33 B0LB15 A0A075X2I7 A0A1I8MHG2 T1PG87 A0A0A1WSG9 Q9U795 A0A0A1XQS3 A0A3G1ZLB6 A0A0J9RFT5 U5EZL1 A0A1L8EFQ5 A0A2A4JII4 K9LHE2 A0A2H1WRE6 A0A1I8Q311 A0A075X2V3 T1PFR3 A0A1W4UHU1 A0A3G1ZLC0 A0A3G1ZLA7 B4HNN4 A0A1L8D6F5 A0A2H1WIN4 A0A1I8MAH9 B4P4L9 A0A0L0CBD3 B4KTI4 A0A1P8PEU4 A0A1W4UG55 A0A1I8MXR0 A0A1I8M6H7 A0A125S6Q2 B4QCE3 A0A1A9WI52 B4QCE4 A0A182WH79 A0A034WM97
Pubmed
EMBL
BABH01008428
BABH01008429
MH177596
AYM01168.1
KF595078
AHB18378.1
+ More
ODYU01007862 SOQ51006.1 MH177581 AYM01153.1 GQ131803 ACZ73898.1 KJ468616 AIB07717.1 AGBW02008896 OWR52256.1 SOQ51004.1 KX778609 AON96571.1 AON96572.1 KF482955 AIH07579.1 NWSH01004968 PCG64545.1 KJ468615 AIB07716.1 MG992377 AXY94815.1 KF929207 AHW45905.1 MH177583 AYM01155.1 SOQ51007.1 SOQ51008.1 GCVX01000118 JAI18112.1 GQ131804 ACZ73899.1 MH177582 AYM01154.1 KM433690 AIZ46902.1 NWSH01006326 PCG63509.1 KQ458793 KPJ05021.1 AGBW02009555 OWR50525.1 KQ459984 KPJ19076.1 KX500034 ARM39004.1 AGBW02010569 OWR48264.1 GCVX01000120 JAI18110.1 GAMC01008536 JAB98019.1 AF526172 AAP75792.1 AF526170 AAP75790.1 KA646899 AFP61528.1 AF526171 AAP75791.1 EU289223 GU170399 ABX84142.1 ADP76648.1 KX500033 ARM39003.1 GAKP01007533 JAC51419.1 GFDG01001368 JAV17431.1 CH933807 EDW12554.1 GFDG01001369 JAV17430.1 AF147205 AAD54937.1 GDHF01033123 GDHF01028856 GDHF01021312 GDHF01020234 GDHF01015969 GDHF01013489 GDHF01013102 GDHF01006194 JAI19191.1 JAI23458.1 JAI31002.1 JAI32080.1 JAI36345.1 JAI38825.1 JAI39212.1 JAI46120.1 KJ468613 AIB07714.1 GDHF01000006 JAI52308.1 BABH01040727 KT852953 ALR34895.1 EF506488 ABS56976.1 KF482960 AIH07584.1 KA646898 AFP61527.1 GBXI01012944 JAD01348.1 AF147206 AAD54938.1 GBXI01017180 GBXI01001379 JAC97111.1 JAD12913.1 MH177589 AYM01161.1 CM002911 KMY94817.1 GANO01000594 JAB59277.1 GFDG01001370 JAV17429.1 NWSH01001367 PCG71518.1 JN003589 AFI99081.1 ODYU01010121 SOQ55004.1 KF482956 AIH07580.1 KA647509 AFP62138.1 MH177588 AYM01160.1 MH177584 AYM01156.1 CH480816 EDW48453.1 GEYN01000101 JAV02028.1 ODYU01008927 SOQ52931.1 CM000158 EDW91642.1 JRES01000655 KNC29542.1 CH933808 EDW08545.1 KU522309 APX61028.1 KT601144 AME15813.1 CM000362 EDX07667.1 KMY94820.1 EDX07668.1 KMY94821.1 GAKP01003198 JAC55754.1
ODYU01007862 SOQ51006.1 MH177581 AYM01153.1 GQ131803 ACZ73898.1 KJ468616 AIB07717.1 AGBW02008896 OWR52256.1 SOQ51004.1 KX778609 AON96571.1 AON96572.1 KF482955 AIH07579.1 NWSH01004968 PCG64545.1 KJ468615 AIB07716.1 MG992377 AXY94815.1 KF929207 AHW45905.1 MH177583 AYM01155.1 SOQ51007.1 SOQ51008.1 GCVX01000118 JAI18112.1 GQ131804 ACZ73899.1 MH177582 AYM01154.1 KM433690 AIZ46902.1 NWSH01006326 PCG63509.1 KQ458793 KPJ05021.1 AGBW02009555 OWR50525.1 KQ459984 KPJ19076.1 KX500034 ARM39004.1 AGBW02010569 OWR48264.1 GCVX01000120 JAI18110.1 GAMC01008536 JAB98019.1 AF526172 AAP75792.1 AF526170 AAP75790.1 KA646899 AFP61528.1 AF526171 AAP75791.1 EU289223 GU170399 ABX84142.1 ADP76648.1 KX500033 ARM39003.1 GAKP01007533 JAC51419.1 GFDG01001368 JAV17431.1 CH933807 EDW12554.1 GFDG01001369 JAV17430.1 AF147205 AAD54937.1 GDHF01033123 GDHF01028856 GDHF01021312 GDHF01020234 GDHF01015969 GDHF01013489 GDHF01013102 GDHF01006194 JAI19191.1 JAI23458.1 JAI31002.1 JAI32080.1 JAI36345.1 JAI38825.1 JAI39212.1 JAI46120.1 KJ468613 AIB07714.1 GDHF01000006 JAI52308.1 BABH01040727 KT852953 ALR34895.1 EF506488 ABS56976.1 KF482960 AIH07584.1 KA646898 AFP61527.1 GBXI01012944 JAD01348.1 AF147206 AAD54938.1 GBXI01017180 GBXI01001379 JAC97111.1 JAD12913.1 MH177589 AYM01161.1 CM002911 KMY94817.1 GANO01000594 JAB59277.1 GFDG01001370 JAV17429.1 NWSH01001367 PCG71518.1 JN003589 AFI99081.1 ODYU01010121 SOQ55004.1 KF482956 AIH07580.1 KA647509 AFP62138.1 MH177588 AYM01160.1 MH177584 AYM01156.1 CH480816 EDW48453.1 GEYN01000101 JAV02028.1 ODYU01008927 SOQ52931.1 CM000158 EDW91642.1 JRES01000655 KNC29542.1 CH933808 EDW08545.1 KU522309 APX61028.1 KT601144 AME15813.1 CM000362 EDX07667.1 KMY94820.1 EDX07668.1 KMY94821.1 GAKP01003198 JAC55754.1
Proteomes
Pfam
Interpro
IPR010987
Glutathione-S-Trfase_C-like
+ More
IPR004046 GST_C
IPR036282 Glutathione-S-Trfase_C_sf
IPR004045 Glutathione_S-Trfase_N
IPR040079 Glutathione_S-Trfase
IPR036249 Thioredoxin-like_sf
IPR027302 Gln_synth_N_conserv_site
IPR014746 Gln_synth/guanido_kin_cat_dom
IPR005574 RPB4/RPC9
IPR010997 HRDC-like_sf
IPR008146 Gln_synth_cat_dom
IPR038324 Rpb4/RPC9_sf
IPR008147 Gln_synt_b-grasp
IPR006590 RNA_pol_Rpb4/RPC9_core
IPR036651 Gln_synt_N
IPR004046 GST_C
IPR036282 Glutathione-S-Trfase_C_sf
IPR004045 Glutathione_S-Trfase_N
IPR040079 Glutathione_S-Trfase
IPR036249 Thioredoxin-like_sf
IPR027302 Gln_synth_N_conserv_site
IPR014746 Gln_synth/guanido_kin_cat_dom
IPR005574 RPB4/RPC9
IPR010997 HRDC-like_sf
IPR008146 Gln_synth_cat_dom
IPR038324 Rpb4/RPC9_sf
IPR008147 Gln_synt_b-grasp
IPR006590 RNA_pol_Rpb4/RPC9_core
IPR036651 Gln_synt_N
SUPFAM
Gene 3D
ProteinModelPortal
H9JAU4
A0A3G1ZLC2
V5QP65
A0A2H1WDG8
A0A3G1ZLA8
D2I930
+ More
A0A0D3M5U3 A0A212FES8 A0A2H1WEV2 A0A1C9EGF0 A0A1C9EGF2 A0A075X2I2 A0A2A4IXK8 A0A0D3M5X3 A0A3G1T1G1 X5CCB3 A0A3G1ZLA9 A0A2H1WDA0 A0A2H1WD57 A0A0K8TUF8 D2I931 A0A3G1ZLB1 A0A0A7KL70 A0A2A4IV17 A0A194QNF8 A0A212F9V3 A0A194RSN3 A0A1W6I4K8 A0A212F3G0 A0A0K8TVL2 W8C6H4 Q7Z0Q7 Q7Z0Q9 T1PDX5 A0A1I8M6Z2 Q7Z0Q8 A9UIB0 A0A1W6I4K6 A0A034W9N4 A0A1L8EFG7 B4KJV1 A0A1L8EFD4 Q9U796 A0A0K8W5E9 A0A0D3M5V9 A0A0K8WMC3 H9JKA0 A0A0S3G4L2 A0A1I8PF33 B0LB15 A0A075X2I7 A0A1I8MHG2 T1PG87 A0A0A1WSG9 Q9U795 A0A0A1XQS3 A0A3G1ZLB6 A0A0J9RFT5 U5EZL1 A0A1L8EFQ5 A0A2A4JII4 K9LHE2 A0A2H1WRE6 A0A1I8Q311 A0A075X2V3 T1PFR3 A0A1W4UHU1 A0A3G1ZLC0 A0A3G1ZLA7 B4HNN4 A0A1L8D6F5 A0A2H1WIN4 A0A1I8MAH9 B4P4L9 A0A0L0CBD3 B4KTI4 A0A1P8PEU4 A0A1W4UG55 A0A1I8MXR0 A0A1I8M6H7 A0A125S6Q2 B4QCE3 A0A1A9WI52 B4QCE4 A0A182WH79 A0A034WM97
A0A0D3M5U3 A0A212FES8 A0A2H1WEV2 A0A1C9EGF0 A0A1C9EGF2 A0A075X2I2 A0A2A4IXK8 A0A0D3M5X3 A0A3G1T1G1 X5CCB3 A0A3G1ZLA9 A0A2H1WDA0 A0A2H1WD57 A0A0K8TUF8 D2I931 A0A3G1ZLB1 A0A0A7KL70 A0A2A4IV17 A0A194QNF8 A0A212F9V3 A0A194RSN3 A0A1W6I4K8 A0A212F3G0 A0A0K8TVL2 W8C6H4 Q7Z0Q7 Q7Z0Q9 T1PDX5 A0A1I8M6Z2 Q7Z0Q8 A9UIB0 A0A1W6I4K6 A0A034W9N4 A0A1L8EFG7 B4KJV1 A0A1L8EFD4 Q9U796 A0A0K8W5E9 A0A0D3M5V9 A0A0K8WMC3 H9JKA0 A0A0S3G4L2 A0A1I8PF33 B0LB15 A0A075X2I7 A0A1I8MHG2 T1PG87 A0A0A1WSG9 Q9U795 A0A0A1XQS3 A0A3G1ZLB6 A0A0J9RFT5 U5EZL1 A0A1L8EFQ5 A0A2A4JII4 K9LHE2 A0A2H1WRE6 A0A1I8Q311 A0A075X2V3 T1PFR3 A0A1W4UHU1 A0A3G1ZLC0 A0A3G1ZLA7 B4HNN4 A0A1L8D6F5 A0A2H1WIN4 A0A1I8MAH9 B4P4L9 A0A0L0CBD3 B4KTI4 A0A1P8PEU4 A0A1W4UG55 A0A1I8MXR0 A0A1I8M6H7 A0A125S6Q2 B4QCE3 A0A1A9WI52 B4QCE4 A0A182WH79 A0A034WM97
PDB
3VWX
E-value=2.03986e-10,
Score=151
Ontologies
GO
Topology
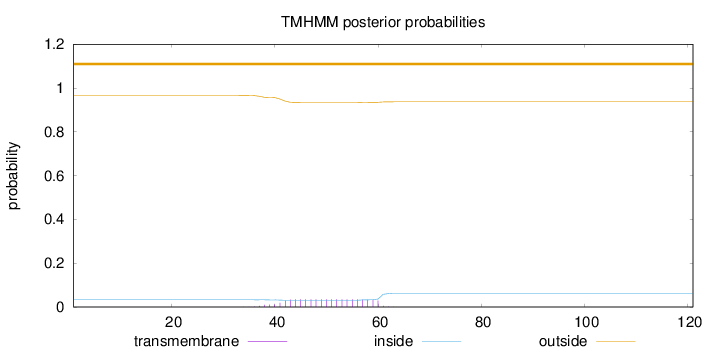
Length:
121
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.71338
Exp number, first 60 AAs:
0.70555
Total prob of N-in:
0.03315
outside
1 - 121
Population Genetic Test Statistics
Pi
142.897233
Theta
115.241556
Tajima's D
-0.661063
CLR
141.203425
CSRT
0.208989550522474
Interpretation
Uncertain
