Gene
KWMTBOMO05455
Pre Gene Modal
BGIBMGA006701
Annotation
PREDICTED:_histone_transcription_regulator_slm9_[Papilio_xuthus]
Full name
Protein HIRA
Location in the cell
Extracellular Reliability : 1.535 Nuclear Reliability : 1.395
Sequence
CDS
ATGAAGCTATTAAAACCCAGTTGGGTCAATCACGATGACAAACCGATATTTTCTGTGGATATACATCCGACTGGAAAATATTTTGCGACCGGGGGACAAGGTGGATACTCGGGCAGAGTAGTTGTGTGGAACTTGAACTCGGTGCTATGTGAATCCGTAGAATTAGACCCAAATGTTCCAAAAATAATTTGCCAAATGGACAACCACCTAGCATGTGTAAATAGTGTCCGTTGGAGTAATGGGGGTAAATACCTGGCTTCTGGAGGAGATGACAAATTGATTATGGTGTGGGGTATTAGTATGGCCCCTGGAGCACCTGGAAAGCACAAAGCTGAAACCTGGAGGTGTTTGTCTACTTTAAGAGGACATGCAGGTGATGTCTTAGACCTAGCATGGTCGCCGTTAGACAAATGGCTGGCTAGCTGCAGCGTGGATAATACAATCATTATATGGAATGCTGAGAAATTTCCAGAAATGGTGTCAGTATTAAATGGTCACACTGGCTTAGTCAAGGGTGTTACATGGGATCCTGTTGGCAAATATTTGGCATCGCAATCTGATGACAAAACCCTGAGGCTATGGAAGACTACTGATTGGGCACAGGAGGCACTCATCACAAAACCATTTTATGAATGTGGGGGCACAACCCACGTTCTTCGTCTTTCCTGGTCTCCGGACGGCCAGTACGTCTTGTCAGCTCACGCGATGAATCGTGGCGGTCCCACTGCGAGGGTCGTGGAGCGAGATGGCTGGAGATGTGATAAAGATTTCGTGGGTCACAGAAAAGCTGTAACTTGTGCTCGCTTTAACAGCAACATCTTCGTGAGGGAAGGCAAGAAGTGCTGTTGTGCCGCCGTCGGGTCCCGGGACCGCGCGCTCAGCATCTGGCAGACGTCGCTGAAGAGGCCGCTGGTCGTCATACACGACCTGTTCAGTGACAGCGTCCTGGATATATCTTGGAGCTCCGATGGGCTCAGCCTTTTAGCTTGTAGTTCGGATGGAACGGTCGCTTGCCTACAGTTTACGAACAAAGAAATCGGTACACCACTCACTCTGGAGGAAAAGAACGCATTCTACGAGAAGATCTATGGTAAAGTTTTGGCGAACGAGTCCGGCGGCGATCTCTCGTCGAACTTACTGGTAGAGTGCCCGGAGATCCTGCTCGCCAGAGAGAAACAGGAAGCCAAGAAGGAAGCTGCGGCAGCTGCTAAGGAAGATTCCACGCCGAAGAGCGAGAAATCACAAAGCGCGCCACTGGCGAGGTCGTCGCCCGCGTCCCCCGGCCCGCCGCGCCCGCTGCGGCCCCTGGACCGACAGATCGAGACGCGCACGTCCGACGGGAAGAGACGCATCACGCCCGTCTTCATACCGCACTCGCAGCTCGACGCCAATGAATCCCTCGGCTCTCAGCCCGGCGGCTCGGAGAACTGTTTCAGCACATCCTCGCAAAGCAAGTCCAGGATCCGGGTGGAAGTCCGGGACGATATCGTGATACACCCGAACGTCAGCAACCACTCCATGTCCGCGCTGCAGAACAACTCTAGGACCTCAGATAACGGTCTAGACAGTCGGCTGCGCAAGCGCACGGCGTCGTCGCTGCCCGGGCCCCGCACCGACGAGCTGCTGCACAAGCGGCCGTGCCCCTCCCCCGCGCCCGCGCCCGCCGCCGCGCCGCTGCTGCCCGCGCCCGCGCTCGGCGTGGCCGGCCCCGTCGTCAGAGCTGCTGGGCCACTGAGGCTGCAGGTGGTGAACAAAGCCTGCAACACGCATTTCGGCACCCTGTCCCGGCTGCAACTCATACCCAACAACTCCGCCTCAAATGATCCCGTTTGGGAAACTTATCTAGGCTCCCCGGTGTGCTGCATCGCGTGCGACGCTCGCTGGGCGTGCGTGTCGTGCGCGGACGGCACGCTGCACGCGTGGCGCGTCTCGCGGTCGCACGCCGCGCGGGCCCTGCCGCCGCTCGCGCTGCCGGCGGCGGCGGCGCGCCTCACGCTGTGTGGGGACACGCTGGCGGCGGTGACGGTGACGGCGGAGCTGGCCATCTGGGACCTGGCCGCCGCCGCCTGCCTCGTGCGACCGCTCTGCTTCAGATCCTTGCTGACTCCTGGAGCTACGGTAACGAATTGTTCTCTACTGGAGGACGGCAACCCTTTGATCTCATTAAGCAACGGGAAGACTTACATATACAGCCAGAAACTGTGCGCCTGGTGA
Protein
MKLLKPSWVNHDDKPIFSVDIHPTGKYFATGGQGGYSGRVVVWNLNSVLCESVELDPNVPKIICQMDNHLACVNSVRWSNGGKYLASGGDDKLIMVWGISMAPGAPGKHKAETWRCLSTLRGHAGDVLDLAWSPLDKWLASCSVDNTIIIWNAEKFPEMVSVLNGHTGLVKGVTWDPVGKYLASQSDDKTLRLWKTTDWAQEALITKPFYECGGTTHVLRLSWSPDGQYVLSAHAMNRGGPTARVVERDGWRCDKDFVGHRKAVTCARFNSNIFVREGKKCCCAAVGSRDRALSIWQTSLKRPLVVIHDLFSDSVLDISWSSDGLSLLACSSDGTVACLQFTNKEIGTPLTLEEKNAFYEKIYGKVLANESGGDLSSNLLVECPEILLAREKQEAKKEAAAAAKEDSTPKSEKSQSAPLARSSPASPGPPRPLRPLDRQIETRTSDGKRRITPVFIPHSQLDANESLGSQPGGSENCFSTSSQSKSRIRVEVRDDIVIHPNVSNHSMSALQNNSRTSDNGLDSRLRKRTASSLPGPRTDELLHKRPCPSPAPAPAAAPLLPAPALGVAGPVVRAAGPLRLQVVNKACNTHFGTLSRLQLIPNNSASNDPVWETYLGSPVCCIACDARWACVSCADGTLHAWRVSRSHAARALPPLALPAAAARLTLCGDTLAAVTVTAELAIWDLAAAACLVRPLCFRSLLTPGATVTNCSLLEDGNPLISLSNGKTYIYSQKLCAW
Summary
Description
Required for replication-independent chromatin assembly and for the periodic repression of histone gene transcription during the cell cycle.
Similarity
Belongs to the WD repeat HIR1 family.
Uniprot
H9JB06
A0A1C9EGF7
A0A194QJB6
A0A2A4IZK4
A0A2J7PX46
A0A2J7PX32
+ More
A0A067RP77 A0A182VUX0 A0A182QHA6 A0A182M5Q8 A0A084VPJ5 A0A182NEH0 A0A182PA84 A0A182IV44 W5JB93 A0A182JSZ8 A0A182F570 A0A182X0F3 A0A182VNK0 A0A182RGD1 A0A026WE51 Q7QG42 A0A2M4AGS2 A0A182TP39 A0A182IE67 A0A182L5J4 A0A1Y1K5P0 A0A1Y1K0F1 A0A1Y1K3B7 A0A1Y1K0P6 A0A1Y1K0G8 E9G6A3 A0A1B6BYQ8 E2BJI9 A0A1I8MYG2 A0A1Q3EY99 A0A0C9R844 A0A182G0Q1 A0A1J1IV56 A0A158NUT4 A0A195CPH3 A0A151K133 A0A182G1C2 A0A151IRA5 A0A1I8PAL3 E2AMT4 A0A1W4XJ25 A0A2M4BCN6 A0A2M4BCP0 W8BYC3 A0A2A3E7T5 A0A0A1WMF7 A0A088AJC3 B0W5Q4 Q0IF51 A0A154PIC9 A0A1B0CV75 H3C2W1 A0A131ZDY3 A0A1Z5L8I4 A0A3B4FN31 H3DH56
A0A067RP77 A0A182VUX0 A0A182QHA6 A0A182M5Q8 A0A084VPJ5 A0A182NEH0 A0A182PA84 A0A182IV44 W5JB93 A0A182JSZ8 A0A182F570 A0A182X0F3 A0A182VNK0 A0A182RGD1 A0A026WE51 Q7QG42 A0A2M4AGS2 A0A182TP39 A0A182IE67 A0A182L5J4 A0A1Y1K5P0 A0A1Y1K0F1 A0A1Y1K3B7 A0A1Y1K0P6 A0A1Y1K0G8 E9G6A3 A0A1B6BYQ8 E2BJI9 A0A1I8MYG2 A0A1Q3EY99 A0A0C9R844 A0A182G0Q1 A0A1J1IV56 A0A158NUT4 A0A195CPH3 A0A151K133 A0A182G1C2 A0A151IRA5 A0A1I8PAL3 E2AMT4 A0A1W4XJ25 A0A2M4BCN6 A0A2M4BCP0 W8BYC3 A0A2A3E7T5 A0A0A1WMF7 A0A088AJC3 B0W5Q4 Q0IF51 A0A154PIC9 A0A1B0CV75 H3C2W1 A0A131ZDY3 A0A1Z5L8I4 A0A3B4FN31 H3DH56
Pubmed
EMBL
BABH01008420
KX778609
AON96565.1
KQ458793
KPJ05015.1
NWSH01004602
+ More
PCG64858.1 NEVH01020862 PNF20894.1 PNF20893.1 KK852507 KDR22430.1 AXCN02000858 AXCM01000440 ATLV01015007 KE524999 KFB39889.1 ADMH02001881 ETN60673.1 KK107261 QOIP01000012 EZA53951.1 RLU15788.1 AAAB01008839 EAA05842.4 GGFK01006672 MBW39993.1 APCN01004570 GEZM01096318 JAV54995.1 GEZM01096321 JAV54992.1 GEZM01096322 GEZM01096317 JAV54991.1 GEZM01096320 JAV54993.1 GEZM01096315 JAV54999.1 GL732533 EFX84898.1 GEDC01030895 GEDC01030892 JAS06403.1 JAS06406.1 GL448571 EFN84146.1 GFDL01014847 JAV20198.1 GBYB01004205 JAG73972.1 JXUM01037136 JXUM01037137 KQ561118 KXJ79650.1 CVRI01000061 CRL04139.1 ADTU01026698 KQ977481 KYN02392.1 KQ981296 KYN43106.1 JXUM01038252 KQ561158 KXJ79512.1 KQ981144 KYN09146.1 GL440896 EFN65248.1 GGFJ01001673 MBW50814.1 GGFJ01001674 MBW50815.1 GAMC01004942 GAMC01004938 JAC01614.1 KZ288338 PBC27807.1 GBXI01014250 JAD00042.1 DS231844 EDS35707.1 CH477394 EAT41888.1 KQ434899 KZC10950.1 AJWK01030162 GEDV01000106 JAP88451.1 GFJQ02003355 JAW03615.1
PCG64858.1 NEVH01020862 PNF20894.1 PNF20893.1 KK852507 KDR22430.1 AXCN02000858 AXCM01000440 ATLV01015007 KE524999 KFB39889.1 ADMH02001881 ETN60673.1 KK107261 QOIP01000012 EZA53951.1 RLU15788.1 AAAB01008839 EAA05842.4 GGFK01006672 MBW39993.1 APCN01004570 GEZM01096318 JAV54995.1 GEZM01096321 JAV54992.1 GEZM01096322 GEZM01096317 JAV54991.1 GEZM01096320 JAV54993.1 GEZM01096315 JAV54999.1 GL732533 EFX84898.1 GEDC01030895 GEDC01030892 JAS06403.1 JAS06406.1 GL448571 EFN84146.1 GFDL01014847 JAV20198.1 GBYB01004205 JAG73972.1 JXUM01037136 JXUM01037137 KQ561118 KXJ79650.1 CVRI01000061 CRL04139.1 ADTU01026698 KQ977481 KYN02392.1 KQ981296 KYN43106.1 JXUM01038252 KQ561158 KXJ79512.1 KQ981144 KYN09146.1 GL440896 EFN65248.1 GGFJ01001673 MBW50814.1 GGFJ01001674 MBW50815.1 GAMC01004942 GAMC01004938 JAC01614.1 KZ288338 PBC27807.1 GBXI01014250 JAD00042.1 DS231844 EDS35707.1 CH477394 EAT41888.1 KQ434899 KZC10950.1 AJWK01030162 GEDV01000106 JAP88451.1 GFJQ02003355 JAW03615.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000235965
UP000027135
UP000075920
+ More
UP000075886 UP000075883 UP000030765 UP000075884 UP000075885 UP000075880 UP000000673 UP000075881 UP000069272 UP000076407 UP000075903 UP000075900 UP000053097 UP000279307 UP000007062 UP000075902 UP000075840 UP000075882 UP000000305 UP000008237 UP000095301 UP000069940 UP000249989 UP000183832 UP000005205 UP000078542 UP000078541 UP000078492 UP000095300 UP000000311 UP000192223 UP000242457 UP000005203 UP000002320 UP000008820 UP000076502 UP000092461 UP000007303 UP000261460
UP000075886 UP000075883 UP000030765 UP000075884 UP000075885 UP000075880 UP000000673 UP000075881 UP000069272 UP000076407 UP000075903 UP000075900 UP000053097 UP000279307 UP000007062 UP000075902 UP000075840 UP000075882 UP000000305 UP000008237 UP000095301 UP000069940 UP000249989 UP000183832 UP000005205 UP000078542 UP000078541 UP000078492 UP000095300 UP000000311 UP000192223 UP000242457 UP000005203 UP000002320 UP000008820 UP000076502 UP000092461 UP000007303 UP000261460
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JB06
A0A1C9EGF7
A0A194QJB6
A0A2A4IZK4
A0A2J7PX46
A0A2J7PX32
+ More
A0A067RP77 A0A182VUX0 A0A182QHA6 A0A182M5Q8 A0A084VPJ5 A0A182NEH0 A0A182PA84 A0A182IV44 W5JB93 A0A182JSZ8 A0A182F570 A0A182X0F3 A0A182VNK0 A0A182RGD1 A0A026WE51 Q7QG42 A0A2M4AGS2 A0A182TP39 A0A182IE67 A0A182L5J4 A0A1Y1K5P0 A0A1Y1K0F1 A0A1Y1K3B7 A0A1Y1K0P6 A0A1Y1K0G8 E9G6A3 A0A1B6BYQ8 E2BJI9 A0A1I8MYG2 A0A1Q3EY99 A0A0C9R844 A0A182G0Q1 A0A1J1IV56 A0A158NUT4 A0A195CPH3 A0A151K133 A0A182G1C2 A0A151IRA5 A0A1I8PAL3 E2AMT4 A0A1W4XJ25 A0A2M4BCN6 A0A2M4BCP0 W8BYC3 A0A2A3E7T5 A0A0A1WMF7 A0A088AJC3 B0W5Q4 Q0IF51 A0A154PIC9 A0A1B0CV75 H3C2W1 A0A131ZDY3 A0A1Z5L8I4 A0A3B4FN31 H3DH56
A0A067RP77 A0A182VUX0 A0A182QHA6 A0A182M5Q8 A0A084VPJ5 A0A182NEH0 A0A182PA84 A0A182IV44 W5JB93 A0A182JSZ8 A0A182F570 A0A182X0F3 A0A182VNK0 A0A182RGD1 A0A026WE51 Q7QG42 A0A2M4AGS2 A0A182TP39 A0A182IE67 A0A182L5J4 A0A1Y1K5P0 A0A1Y1K0F1 A0A1Y1K3B7 A0A1Y1K0P6 A0A1Y1K0G8 E9G6A3 A0A1B6BYQ8 E2BJI9 A0A1I8MYG2 A0A1Q3EY99 A0A0C9R844 A0A182G0Q1 A0A1J1IV56 A0A158NUT4 A0A195CPH3 A0A151K133 A0A182G1C2 A0A151IRA5 A0A1I8PAL3 E2AMT4 A0A1W4XJ25 A0A2M4BCN6 A0A2M4BCP0 W8BYC3 A0A2A3E7T5 A0A0A1WMF7 A0A088AJC3 B0W5Q4 Q0IF51 A0A154PIC9 A0A1B0CV75 H3C2W1 A0A131ZDY3 A0A1Z5L8I4 A0A3B4FN31 H3DH56
PDB
2YMU
E-value=1.66029e-17,
Score=222
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
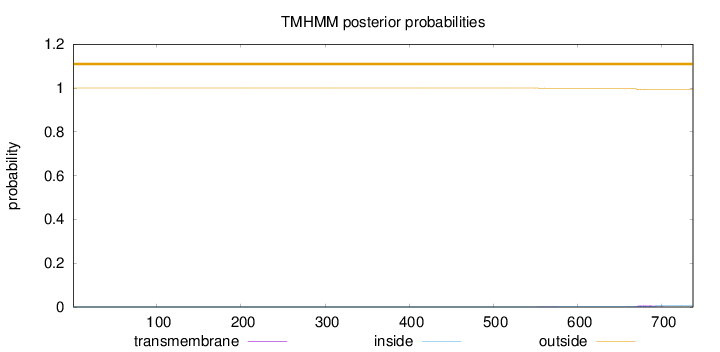
Length:
737
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1666
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00012
outside
1 - 737
Population Genetic Test Statistics
Pi
252.723808
Theta
175.865977
Tajima's D
0.109186
CLR
16.280331
CSRT
0.393730313484326
Interpretation
Uncertain
