Gene
KWMTBOMO05453
Annotation
PREDICTED:_solute_carrier_family_2?_facilitated_glucose_transporter_member_1-like_[Bombyx_mori]
Full name
Solute carrier family 2, facilitated glucose transporter member 4
Alternative Name
Glucose transporter type 4, insulin-responsive
Location in the cell
PlasmaMembrane Reliability : 4.946
Sequence
CDS
ATGGGGGACAGAGCAGCTGGTTCTACCCTGCTTGCCGAAAATCGGCGACGAGAATCTTTTCAATATGTACCACAACCGGTATTCAATGAGAACCAGCATTGGACTAAGACGAACGAGTACACAGCCACCGGGTGGTCGTTTTATTTGATATTGGCCGGAGTCATCACCACCATCGGATCATCCGTCCCTGTAGGATATAATATAGGTGTCGTTAATACACCGGCTGAGGTGATAAAACAATTCTGTAATGAAAGTTTCATCAGCCGTTACGATGTAGTGCTGGACGAGCAATGGATGAATGTTCTTTGGTCTTCGGTGGTGTCTATCTTCATCGTCGGCGGGTGTACCGGCTCTATACTTGGCTCGTACTTAGCTGATAGCCTCGGAAGAAAAATGGCAACGATCGCTACGGGGGCGCTATGGATCGCGGGAGCTTTGCTCTTCTTGCTATGCCGGACGGCTAATTCAGTGGAGATGCTTATAATCGGGAGACTTTTAGTCGGTCTAGCCGGAGGTCTGACTACCAGCATAGTTCCCATGTACCTGACGGAGCTGGCGCCGCCCAGACTAACTGGAGCCATGGGTGTCGCGTGTCCCATGGGGGTCAACGTGGGTGTTCTTGTTGGGCAAGTTATGGGATTGAACTTTTTGTTGGGGGGCCCGGACAACTGGCCGTATCTGCTAGGGGCTTACGCGATTCTAGTCATTGTGTGTTTACCCATACTGTTCATATTGCCGGAGAGCCCGAAGTACTTGTACGTTATGAAACGCGACGAAGGAAAAGCTTTGGAAGAGTTGAGTCGTCTCCGCGGTGTTTCAATGAGCCTGCTGGCGTCAGAGCTGGAGACTCTTCGTGAGGAAGCGGCCACGGCGTCCGTCGTCCAGCGCTACTCGATGCTGCAGGTGGCGCGGGAGCCTGCCCTGCGCCTGCCGCTGCTCCTGGTGGTGGCGCTACAAGCGGGACAGCAGACCAGCGGGATCAACGCGGTGTTCTACTACTCGCAAATGATGTTCGAGCAGGCCGGTCTGACGGCGTACCAGGCTCAGCTGGCGACGATAGCGGCGGGACTCGTGAACGTGTGCACAGCCGCGCTCATGGTGCGCCTCCTGCCGCGCGCCGGCCGCCGCCCCCTCGTGCTGCGCTCCGTGCTCGCCGCCGCCCTCCTCCTGGCCGCCCTGGCCGCCGCCCTTAACTACATGGACGCGGCGCCCTGGATGCCGTACGTGTGCATAGTGGCCGTGTTGTCTTACGTCAACGCGTACGGGTTCGGTCTTGGACCTATAGCGTATTTTATTGCGTCCGAAATGTTCGAGGTGGCGCCGCGCCCCGCCGCCATGGCGTGGGGCTCGTTGGCGAACTGGGGGGGGAACTTTCTCATCGGAATGTCCTTCCAAACGATGCGGGACACCATCGGCGCCTACTCCTTCTTGTTGTTCTCCGCCGTTACCGCAGCCCTGTACGTTTTTATGAGACTGTACTTCCCCGAAACACGAGGAAAGACTCCGGCTCAAGTATCACTCCTATGCAGCCAAGGGTTCCGCTCCAGACCCCTCAGTACATCACAATTGCGTAATGCTGACGTATGA
Protein
MGDRAAGSTLLAENRRRESFQYVPQPVFNENQHWTKTNEYTATGWSFYLILAGVITTIGSSVPVGYNIGVVNTPAEVIKQFCNESFISRYDVVLDEQWMNVLWSSVVSIFIVGGCTGSILGSYLADSLGRKMATIATGALWIAGALLFLLCRTANSVEMLIIGRLLVGLAGGLTTSIVPMYLTELAPPRLTGAMGVACPMGVNVGVLVGQVMGLNFLLGGPDNWPYLLGAYAILVIVCLPILFILPESPKYLYVMKRDEGKALEELSRLRGVSMSLLASELETLREEAATASVVQRYSMLQVAREPALRLPLLLVVALQAGQQTSGINAVFYYSQMMFEQAGLTAYQAQLATIAAGLVNVCTAALMVRLLPRAGRRPLVLRSVLAAALLLAALAAALNYMDAAPWMPYVCIVAVLSYVNAYGFGLGPIAYFIASEMFEVAPRPAAMAWGSLANWGGNFLIGMSFQTMRDTIGAYSFLLFSAVTAALYVFMRLYFPETRGKTPAQVSLLCSQGFRSRPLSTSQLRNADV
Summary
Description
Insulin-regulated facilitative glucose transporter.
Subunit
Binds to DAXX. Interacts via its N-terminus with SRFBP1 (By similarity). Interacts with NDUFA9 (By similarity). Interacts with TRARG1; the interaction is required for proper SLC2A4 reacycling after insulin stimulation (By similarity).
Miscellaneous
Insulin-stimulated phosphorylation of TBC1D4 is required for GLUT4 translocation.
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the glycosyl hydrolase 18 family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Glucose transporter subfamily.
Belongs to the glycosyl hydrolase 18 family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Glucose transporter subfamily.
Keywords
Cell membrane
Complete proteome
Cytoplasm
Glycoprotein
Membrane
Phosphoprotein
Reference proteome
Sugar transport
Transmembrane
Transmembrane helix
Transport
Ubl conjugation
Feature
chain Solute carrier family 2, facilitated glucose transporter member 4
Uniprot
H9JAU7
A0A1C9EGF9
A0A194QHZ3
A0A212FET9
A0A194RP44
A0A0V0GA77
+ More
A0A0P4VVV0 A0A0C9QBB7 T1I7R9 A0A0J7KUY3 E9IGW0 E2AX78 A0A2A3EL56 A0A087ZWD5 A0A232FN99 K7ITD7 A0A0N0U4U0 A0A310SWN2 A0A0L7QV88 A0A151HYI0 A0A151IQL3 F4X4W4 A0A3L8DGY7 A0A0A1X8M8 A0A0K8VSD7 A0A0K8US51 A0A1A9WWK3 W8BB30 A0A034WJ29 A0A1Z1XG36 A0A195ETA1 A0A0A9XPY7 A0A026WRM9 A0A0K8SSB0 E2BWS1 A0A151WL51 B3N3H4 B4QCQ0 B4II19 Q0IGX4 B4NYW7 A0A3B0J3B3 A0A1W4VYD3 B4LJL7 Q28YP4 B4H8C8 A0A0R3NP04 B4LNN0 J9K8D7 A0A154PSD6 I3MBY4 I3N8R3 A0A2U3VTD2 A0A0M3QUC7 G3TCE1 A0A3Q7PV76 A0A158NVY7 K7FZI7 I1W0Z5 I1W0Z6 I1W101 M3Z096 S7NG63 G1P2Z3 I1W109 L5M095 I1W103 A0A286XET5 A0A3G1I4G1 I0BWM0 I1W107 A0A0N8BMS5 D7RFB4 Q27994 I1W108 A0A212DAE3 Q6EAS4 L5JWG4 K9IKS0 A0A340X8V1 A0A315VZ20 Q8MJ15 A0A1B1RV26 L9KR18 I1W0Z9 I1W100 Q8WSW5 A0A0N8EAH6 H2NSI2 A0A340XCK2 A0A2K6ENC4 A0A2K6ENB3 A0A2R5L7U2 A0A164SYQ9 I1W104 W5PP99 A0A096P2N6 A0A0P5C448 A0A2K6CH25
A0A0P4VVV0 A0A0C9QBB7 T1I7R9 A0A0J7KUY3 E9IGW0 E2AX78 A0A2A3EL56 A0A087ZWD5 A0A232FN99 K7ITD7 A0A0N0U4U0 A0A310SWN2 A0A0L7QV88 A0A151HYI0 A0A151IQL3 F4X4W4 A0A3L8DGY7 A0A0A1X8M8 A0A0K8VSD7 A0A0K8US51 A0A1A9WWK3 W8BB30 A0A034WJ29 A0A1Z1XG36 A0A195ETA1 A0A0A9XPY7 A0A026WRM9 A0A0K8SSB0 E2BWS1 A0A151WL51 B3N3H4 B4QCQ0 B4II19 Q0IGX4 B4NYW7 A0A3B0J3B3 A0A1W4VYD3 B4LJL7 Q28YP4 B4H8C8 A0A0R3NP04 B4LNN0 J9K8D7 A0A154PSD6 I3MBY4 I3N8R3 A0A2U3VTD2 A0A0M3QUC7 G3TCE1 A0A3Q7PV76 A0A158NVY7 K7FZI7 I1W0Z5 I1W0Z6 I1W101 M3Z096 S7NG63 G1P2Z3 I1W109 L5M095 I1W103 A0A286XET5 A0A3G1I4G1 I0BWM0 I1W107 A0A0N8BMS5 D7RFB4 Q27994 I1W108 A0A212DAE3 Q6EAS4 L5JWG4 K9IKS0 A0A340X8V1 A0A315VZ20 Q8MJ15 A0A1B1RV26 L9KR18 I1W0Z9 I1W100 Q8WSW5 A0A0N8EAH6 H2NSI2 A0A340XCK2 A0A2K6ENC4 A0A2K6ENB3 A0A2R5L7U2 A0A164SYQ9 I1W104 W5PP99 A0A096P2N6 A0A0P5C448 A0A2K6CH25
Pubmed
19121390
26354079
22118469
27129103
21282665
20798317
+ More
28648823 20075255 21719571 30249741 25830018 24495485 25348373 28588501 25401762 26823975 24508170 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 21347285 17381049 22493665 21993624 28440667 24381100 9027564 8925410 15366377 23258410 29703783 19892987 27427908 23385571 20809919
28648823 20075255 21719571 30249741 25830018 24495485 25348373 28588501 25401762 26823975 24508170 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 21347285 17381049 22493665 21993624 28440667 24381100 9027564 8925410 15366377 23258410 29703783 19892987 27427908 23385571 20809919
EMBL
BABH01008412
BABH01008413
BABH01008414
KX778609
AON96566.1
KQ458793
+ More
KPJ05014.1 AGBW02008897 OWR52252.1 KQ459984 KPJ19080.1 GECL01001163 JAP04961.1 GDKW01002822 JAI53773.1 GBYB01011380 GBYB01011381 GBYB01011723 JAG81147.1 JAG81148.1 JAG81490.1 ACPB03023874 LBMM01002844 KMQ94292.1 GL763054 EFZ20202.1 GL443520 EFN61961.1 KZ288215 PBC32543.1 NNAY01000006 OXU32115.1 AAZX01003312 KQ435821 KOX72424.1 KQ759851 OAD62556.1 KQ414727 KOC62479.1 KQ976730 KYM76416.1 KQ976772 KYN08452.1 GL888679 EGI58537.1 QOIP01000009 RLU19128.1 GBXI01006820 JAD07472.1 GDHF01010566 JAI41748.1 GDHF01022785 JAI29529.1 GAMC01012347 JAB94208.1 GAKP01004605 JAC54347.1 KY350169 ARX98208.1 KQ981993 KYN31109.1 GBHO01024486 GBHO01024481 GBHO01024476 GBHO01024474 GDHC01005425 GDHC01004839 JAG19118.1 JAG19123.1 JAG19128.1 JAG19130.1 JAQ13204.1 JAQ13790.1 KK107119 EZA58672.1 GBRD01010104 JAG55720.1 GL451188 EFN79785.1 KQ982974 KYQ48584.1 CH954177 EDV59856.2 CM000362 CM002911 EDX05839.1 KMY91668.1 CH480841 EDW49545.1 AE013599 BT028796 AAF57300.2 ABI34177.1 CM000157 EDW89818.2 OUUW01000001 SPP75807.1 CH940648 EDW61585.2 CM000071 EAL25921.2 CH479223 EDW34963.1 KRT02674.1 EDW62210.2 ABLF02041154 KQ435127 KZC14812.1 AGTP01070895 AGTP01107584 CP012524 ALC40433.1 ADTU01027553 ADTU01027554 ADTU01027555 ADTU01027556 AGCU01013080 AGCU01013081 AGCU01013082 JN695651 AFI57553.1 JN695652 AFI57554.1 JN695657 AFI57559.1 AEYP01041520 KE164153 EPQ15455.1 AAPE02052135 AAPE02052136 JN695665 AFI57567.1 KB105946 ELK31781.1 JN695659 AFI57561.1 AAKN02044424 KX168424 ARQ15180.1 JQ343218 AFH66818.1 JN695663 AFI57565.1 GDIQ01162459 JAK89266.1 HM055486 ADI33304.1 D63150 AY458600 BC114082 U18105 JN695664 AFI57566.1 MKHE01000005 OWK15203.1 AY505108 AAS91787.1 KB031076 ELK03794.1 GABZ01005737 JAA47788.1 NHOQ01000739 PWA28910.1 AF531753 AAM95953.2 KT377191 ANU06113.1 KB320771 ELW63617.1 JN695655 AFI57557.1 JN695656 AFI57558.1 AY059413 AAL27090.1 GDIQ01045860 JAN48877.1 ABGA01347993 NDHI03003640 PNJ13036.1 GGLE01001423 MBY05549.1 LRGB01001899 KZS10076.1 JN695660 AFI57562.1 AMGL01018020 AMGL01018021 AHZZ02009643 GDIP01175870 JAJ47532.1
KPJ05014.1 AGBW02008897 OWR52252.1 KQ459984 KPJ19080.1 GECL01001163 JAP04961.1 GDKW01002822 JAI53773.1 GBYB01011380 GBYB01011381 GBYB01011723 JAG81147.1 JAG81148.1 JAG81490.1 ACPB03023874 LBMM01002844 KMQ94292.1 GL763054 EFZ20202.1 GL443520 EFN61961.1 KZ288215 PBC32543.1 NNAY01000006 OXU32115.1 AAZX01003312 KQ435821 KOX72424.1 KQ759851 OAD62556.1 KQ414727 KOC62479.1 KQ976730 KYM76416.1 KQ976772 KYN08452.1 GL888679 EGI58537.1 QOIP01000009 RLU19128.1 GBXI01006820 JAD07472.1 GDHF01010566 JAI41748.1 GDHF01022785 JAI29529.1 GAMC01012347 JAB94208.1 GAKP01004605 JAC54347.1 KY350169 ARX98208.1 KQ981993 KYN31109.1 GBHO01024486 GBHO01024481 GBHO01024476 GBHO01024474 GDHC01005425 GDHC01004839 JAG19118.1 JAG19123.1 JAG19128.1 JAG19130.1 JAQ13204.1 JAQ13790.1 KK107119 EZA58672.1 GBRD01010104 JAG55720.1 GL451188 EFN79785.1 KQ982974 KYQ48584.1 CH954177 EDV59856.2 CM000362 CM002911 EDX05839.1 KMY91668.1 CH480841 EDW49545.1 AE013599 BT028796 AAF57300.2 ABI34177.1 CM000157 EDW89818.2 OUUW01000001 SPP75807.1 CH940648 EDW61585.2 CM000071 EAL25921.2 CH479223 EDW34963.1 KRT02674.1 EDW62210.2 ABLF02041154 KQ435127 KZC14812.1 AGTP01070895 AGTP01107584 CP012524 ALC40433.1 ADTU01027553 ADTU01027554 ADTU01027555 ADTU01027556 AGCU01013080 AGCU01013081 AGCU01013082 JN695651 AFI57553.1 JN695652 AFI57554.1 JN695657 AFI57559.1 AEYP01041520 KE164153 EPQ15455.1 AAPE02052135 AAPE02052136 JN695665 AFI57567.1 KB105946 ELK31781.1 JN695659 AFI57561.1 AAKN02044424 KX168424 ARQ15180.1 JQ343218 AFH66818.1 JN695663 AFI57565.1 GDIQ01162459 JAK89266.1 HM055486 ADI33304.1 D63150 AY458600 BC114082 U18105 JN695664 AFI57566.1 MKHE01000005 OWK15203.1 AY505108 AAS91787.1 KB031076 ELK03794.1 GABZ01005737 JAA47788.1 NHOQ01000739 PWA28910.1 AF531753 AAM95953.2 KT377191 ANU06113.1 KB320771 ELW63617.1 JN695655 AFI57557.1 JN695656 AFI57558.1 AY059413 AAL27090.1 GDIQ01045860 JAN48877.1 ABGA01347993 NDHI03003640 PNJ13036.1 GGLE01001423 MBY05549.1 LRGB01001899 KZS10076.1 JN695660 AFI57562.1 AMGL01018020 AMGL01018021 AHZZ02009643 GDIP01175870 JAJ47532.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000015103
UP000036403
+ More
UP000000311 UP000242457 UP000005203 UP000215335 UP000002358 UP000053105 UP000053825 UP000078540 UP000078542 UP000007755 UP000279307 UP000091820 UP000078541 UP000053097 UP000008237 UP000075809 UP000008711 UP000000304 UP000001292 UP000000803 UP000002282 UP000268350 UP000192221 UP000008792 UP000001819 UP000008744 UP000007819 UP000076502 UP000005215 UP000245340 UP000092553 UP000007646 UP000286641 UP000005205 UP000007267 UP000000715 UP000001074 UP000005447 UP000009136 UP000010552 UP000265300 UP000002281 UP000011518 UP000001595 UP000233160 UP000076858 UP000002356 UP000028761 UP000233120
UP000000311 UP000242457 UP000005203 UP000215335 UP000002358 UP000053105 UP000053825 UP000078540 UP000078542 UP000007755 UP000279307 UP000091820 UP000078541 UP000053097 UP000008237 UP000075809 UP000008711 UP000000304 UP000001292 UP000000803 UP000002282 UP000268350 UP000192221 UP000008792 UP000001819 UP000008744 UP000007819 UP000076502 UP000005215 UP000245340 UP000092553 UP000007646 UP000286641 UP000005205 UP000007267 UP000000715 UP000001074 UP000005447 UP000009136 UP000010552 UP000265300 UP000002281 UP000011518 UP000001595 UP000233160 UP000076858 UP000002356 UP000028761 UP000233120
Interpro
IPR036259
MFS_trans_sf
+ More
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR003663 Sugar/inositol_transpt
IPR005828 MFS_sugar_transport-like
IPR011583 Chitinase_II
IPR036508 Chitin-bd_dom_sf
IPR017853 Glycoside_hydrolase_SF
IPR002557 Chitin-bd_dom
IPR001579 Glyco_hydro_18_chit_AS
IPR001223 Glyco_hydro18_cat
IPR029070 Chitinase_insertion_sf
IPR002441 Glc_transpt_4
IPR002945 Glc_transpt_3
IPR002439 Glu_transpt_1
IPR020846 MFS_dom
IPR005829 Sugar_transporter_CS
IPR003663 Sugar/inositol_transpt
IPR005828 MFS_sugar_transport-like
IPR011583 Chitinase_II
IPR036508 Chitin-bd_dom_sf
IPR017853 Glycoside_hydrolase_SF
IPR002557 Chitin-bd_dom
IPR001579 Glyco_hydro_18_chit_AS
IPR001223 Glyco_hydro18_cat
IPR029070 Chitinase_insertion_sf
IPR002441 Glc_transpt_4
IPR002945 Glc_transpt_3
IPR002439 Glu_transpt_1
Gene 3D
CDD
ProteinModelPortal
H9JAU7
A0A1C9EGF9
A0A194QHZ3
A0A212FET9
A0A194RP44
A0A0V0GA77
+ More
A0A0P4VVV0 A0A0C9QBB7 T1I7R9 A0A0J7KUY3 E9IGW0 E2AX78 A0A2A3EL56 A0A087ZWD5 A0A232FN99 K7ITD7 A0A0N0U4U0 A0A310SWN2 A0A0L7QV88 A0A151HYI0 A0A151IQL3 F4X4W4 A0A3L8DGY7 A0A0A1X8M8 A0A0K8VSD7 A0A0K8US51 A0A1A9WWK3 W8BB30 A0A034WJ29 A0A1Z1XG36 A0A195ETA1 A0A0A9XPY7 A0A026WRM9 A0A0K8SSB0 E2BWS1 A0A151WL51 B3N3H4 B4QCQ0 B4II19 Q0IGX4 B4NYW7 A0A3B0J3B3 A0A1W4VYD3 B4LJL7 Q28YP4 B4H8C8 A0A0R3NP04 B4LNN0 J9K8D7 A0A154PSD6 I3MBY4 I3N8R3 A0A2U3VTD2 A0A0M3QUC7 G3TCE1 A0A3Q7PV76 A0A158NVY7 K7FZI7 I1W0Z5 I1W0Z6 I1W101 M3Z096 S7NG63 G1P2Z3 I1W109 L5M095 I1W103 A0A286XET5 A0A3G1I4G1 I0BWM0 I1W107 A0A0N8BMS5 D7RFB4 Q27994 I1W108 A0A212DAE3 Q6EAS4 L5JWG4 K9IKS0 A0A340X8V1 A0A315VZ20 Q8MJ15 A0A1B1RV26 L9KR18 I1W0Z9 I1W100 Q8WSW5 A0A0N8EAH6 H2NSI2 A0A340XCK2 A0A2K6ENC4 A0A2K6ENB3 A0A2R5L7U2 A0A164SYQ9 I1W104 W5PP99 A0A096P2N6 A0A0P5C448 A0A2K6CH25
A0A0P4VVV0 A0A0C9QBB7 T1I7R9 A0A0J7KUY3 E9IGW0 E2AX78 A0A2A3EL56 A0A087ZWD5 A0A232FN99 K7ITD7 A0A0N0U4U0 A0A310SWN2 A0A0L7QV88 A0A151HYI0 A0A151IQL3 F4X4W4 A0A3L8DGY7 A0A0A1X8M8 A0A0K8VSD7 A0A0K8US51 A0A1A9WWK3 W8BB30 A0A034WJ29 A0A1Z1XG36 A0A195ETA1 A0A0A9XPY7 A0A026WRM9 A0A0K8SSB0 E2BWS1 A0A151WL51 B3N3H4 B4QCQ0 B4II19 Q0IGX4 B4NYW7 A0A3B0J3B3 A0A1W4VYD3 B4LJL7 Q28YP4 B4H8C8 A0A0R3NP04 B4LNN0 J9K8D7 A0A154PSD6 I3MBY4 I3N8R3 A0A2U3VTD2 A0A0M3QUC7 G3TCE1 A0A3Q7PV76 A0A158NVY7 K7FZI7 I1W0Z5 I1W0Z6 I1W101 M3Z096 S7NG63 G1P2Z3 I1W109 L5M095 I1W103 A0A286XET5 A0A3G1I4G1 I0BWM0 I1W107 A0A0N8BMS5 D7RFB4 Q27994 I1W108 A0A212DAE3 Q6EAS4 L5JWG4 K9IKS0 A0A340X8V1 A0A315VZ20 Q8MJ15 A0A1B1RV26 L9KR18 I1W0Z9 I1W100 Q8WSW5 A0A0N8EAH6 H2NSI2 A0A340XCK2 A0A2K6ENC4 A0A2K6ENB3 A0A2R5L7U2 A0A164SYQ9 I1W104 W5PP99 A0A096P2N6 A0A0P5C448 A0A2K6CH25
PDB
5EQI
E-value=1.06202e-71,
Score=688
Ontologies
GO
GO:0016021
GO:0022857
GO:0008643
GO:0005576
GO:0005975
GO:0008061
GO:0004553
GO:0006030
GO:0071470
GO:0071356
GO:0042383
GO:0042593
GO:0044381
GO:0050873
GO:0005355
GO:0005829
GO:0048471
GO:0030140
GO:0030659
GO:0010021
GO:0070062
GO:0009897
GO:0045121
GO:0005887
GO:0032593
GO:0005768
GO:0032869
GO:0016324
GO:0005737
GO:0033300
GO:0005903
GO:0055056
GO:0005911
GO:0012505
GO:0005886
GO:0002080
GO:0016020
GO:0022891
GO:0005215
GO:0050660
GO:0016627
GO:0008152
GO:0016491
GO:0003824
GO:0055085
Topology
Subcellular location
Cell membrane
Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
Endomembrane system Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
Cytoplasm Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
Perinuclear region Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
Endomembrane system Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
Cytoplasm Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
Perinuclear region Localizes primarily to the perinuclear region, undergoing continued recycling to the plasma membrane where it is rapidly reinternalized (By similarity). The dileucine internalization motif is critical for intracellular sequestration (By similarity). Insulin stimulation induces translocation to the cell membrane (By similarity). With evidence from 2 publications.
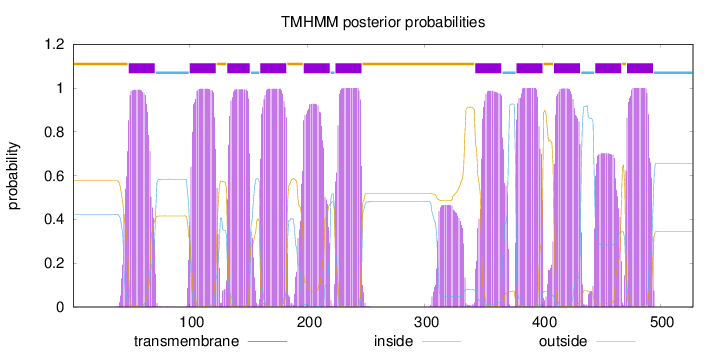
Length:
528
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
244.260020000001
Exp number, first 60 AAs:
15.37759
Total prob of N-in:
0.42347
POSSIBLE N-term signal
sequence
outside
1 - 47
TMhelix
48 - 70
inside
71 - 99
TMhelix
100 - 122
outside
123 - 131
TMhelix
132 - 151
inside
152 - 159
TMhelix
160 - 182
outside
183 - 196
TMhelix
197 - 219
inside
220 - 223
TMhelix
224 - 246
outside
247 - 342
TMhelix
343 - 365
inside
366 - 377
TMhelix
378 - 400
outside
401 - 409
TMhelix
410 - 432
inside
433 - 444
TMhelix
445 - 467
outside
468 - 471
TMhelix
472 - 494
inside
495 - 528
Population Genetic Test Statistics
Pi
239.684056
Theta
202.75164
Tajima's D
0.632895
CLR
0.465674
CSRT
0.553972301384931
Interpretation
Uncertain
