Gene
KWMTBOMO05447
Pre Gene Modal
BGIBMGA006697
Annotation
PREDICTED:_YEATS_domain-containing_protein_4_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 3.31
Sequence
CDS
ATGAGCCTTACGACAGATTTCGGGCCAGATTCTGGCGGCCGTGTGAAGGGTCTAACAATAGTAAAGCCTATAGTTTACGGTAATATAGCCCGTTACTTCGGCAAGAAACGCGAAGAGGATGGACACACACATCAATGGACTGTTTATGTAAAACCTTACGCAAACGAGGATATGTCGACTTACATCAAAAAAGTTCACTTCAAACTTCACGAAAGCTACGCGAACCCCAATAGGATCGTGACCAAGCCCCCTTACGAACTCATGGAGACTGGTTGGGGAGAGTTCGAGATCGTTATAAAGTTATACTTTCATGATCCCAATGAAAGACCACTTACTCTATACCACATACTGAAATTATTCCAATCGCCAGTCACTGAAGGTACTCCCCCAACATTTGGACATGCTCTCGTCAGTGAATCTTATGAGGAGATAGTATTCCAAGAACCAACTCAACTAATGCAGCATCTGTTAAAGAATGTCAGACCAATTACGAATGGACATTGGAAACATGACACGGACTTTGAAGAGAAGAAAGAGAAGACTTTAGAAAGAATAATAGAAGCTCAAACAAAAGTGAGGACAGAGATATCAGAGCTGAAAGAGAAACTTAGTCTTGCAAAGGAAACCATAGCTAAGTTCAAGGAAGAAATTGCCAAACTACAGAATAGTTCAGGGAACAATATAATGATGTAA
Protein
MSLTTDFGPDSGGRVKGLTIVKPIVYGNIARYFGKKREEDGHTHQWTVYVKPYANEDMSTYIKKVHFKLHESYANPNRIVTKPPYELMETGWGEFEIVIKLYFHDPNERPLTLYHILKLFQSPVTEGTPPTFGHALVSESYEEIVFQEPTQLMQHLLKNVRPITNGHWKHDTDFEEKKEKTLERIIEAQTKVRTEISELKEKLSLAKETIAKFKEEIAKLQNSSGNNIMM
Summary
Similarity
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Uniprot
H9JB02
A0A2H1V376
A0A2A4J341
A0A1E1WAA2
A0A194RNK2
A0A1C9EGF5
+ More
A0A194QHI5 A0A3S2NX48 A0A212FEQ9 A0A1Q3EZ26 B0VZZ6 A0A182JFM0 A0A1B0D2C8 A0A1B0CVQ1 A0A0L7QVG5 Q16LU6 A0A182G6R9 A0A1L8DGV8 A0A087ZWB7 A0A182PKV2 A0A2M4BYQ4 A0A1I8NA85 A0A2M4BYS3 A0A026WRK6 A0A1I8NRD6 A0A310SS33 A0A182F4D3 W5J696 A0A0N0U4R3 A0A154PUD2 F4X4W2 A0A195ET69 E9IGW2 A0A151IQL1 A0A158NVW3 A0A182TYY1 A0A182WZD9 A0A182L669 Q7Q1S4 A0A182ICS8 A0A182NAX8 T1GFC6 A0A182Q6F0 A0A182VD91 A0A2A3EMP5 A0A0K8TMY6 A0A182Y8B3 A0A336L1Y9 A0A182M7K7 A0A151WL25 U4UBW2 A0A182VTW6 A0A0K8VJ50 A0A034WKC8 A0A2J7RS70 E2AX80 W8C7R2 A0A182JNR6 A0A0K8UY36 K7IWJ2 D6WZM9 A0A0C9QAC1 A0A1A9WEA3 A0A1B6IJ38 A0A1B6MC32 A0A1B6G4R2 D3TR93 A0A1A9Y1I4 A0A1A9VYS8 A0A067QXQ0 A0A2P8XK76 N6SZK2 T1JBJ9 A0A182RGQ3 A0A0P4VV85 A0A0N7ZAT7 E0VVP5 D3TR94 A0A1B6D9X6 A0A224XXN6 A0A1Y1LRD1 Q29JX5 B4GKZ9 B3MKS2 A0A1W4UBD1 B4Q5A1 A0A2M3ZD09 A0A1B0FB53 Q9VM63 B4HY04 B4NZY7 B4N073 A0A0M4E7C1 R4G419 B4KJW0 B4JEF1 B4LRN6 A0A1W4XLN3 A0A1S3JLX6 A0A0L7LCJ9
A0A194QHI5 A0A3S2NX48 A0A212FEQ9 A0A1Q3EZ26 B0VZZ6 A0A182JFM0 A0A1B0D2C8 A0A1B0CVQ1 A0A0L7QVG5 Q16LU6 A0A182G6R9 A0A1L8DGV8 A0A087ZWB7 A0A182PKV2 A0A2M4BYQ4 A0A1I8NA85 A0A2M4BYS3 A0A026WRK6 A0A1I8NRD6 A0A310SS33 A0A182F4D3 W5J696 A0A0N0U4R3 A0A154PUD2 F4X4W2 A0A195ET69 E9IGW2 A0A151IQL1 A0A158NVW3 A0A182TYY1 A0A182WZD9 A0A182L669 Q7Q1S4 A0A182ICS8 A0A182NAX8 T1GFC6 A0A182Q6F0 A0A182VD91 A0A2A3EMP5 A0A0K8TMY6 A0A182Y8B3 A0A336L1Y9 A0A182M7K7 A0A151WL25 U4UBW2 A0A182VTW6 A0A0K8VJ50 A0A034WKC8 A0A2J7RS70 E2AX80 W8C7R2 A0A182JNR6 A0A0K8UY36 K7IWJ2 D6WZM9 A0A0C9QAC1 A0A1A9WEA3 A0A1B6IJ38 A0A1B6MC32 A0A1B6G4R2 D3TR93 A0A1A9Y1I4 A0A1A9VYS8 A0A067QXQ0 A0A2P8XK76 N6SZK2 T1JBJ9 A0A182RGQ3 A0A0P4VV85 A0A0N7ZAT7 E0VVP5 D3TR94 A0A1B6D9X6 A0A224XXN6 A0A1Y1LRD1 Q29JX5 B4GKZ9 B3MKS2 A0A1W4UBD1 B4Q5A1 A0A2M3ZD09 A0A1B0FB53 Q9VM63 B4HY04 B4NZY7 B4N073 A0A0M4E7C1 R4G419 B4KJW0 B4JEF1 B4LRN6 A0A1W4XLN3 A0A1S3JLX6 A0A0L7LCJ9
Pubmed
19121390
26354079
22118469
17510324
26483478
25315136
+ More
24508170 30249741 20920257 23761445 21719571 21282665 21347285 20966253 12364791 14747013 17210077 26369729 25244985 23537049 25348373 20798317 24495485 20075255 18362917 19820115 20353571 24845553 29403074 20566863 28004739 15632085 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26227816
24508170 30249741 20920257 23761445 21719571 21282665 21347285 20966253 12364791 14747013 17210077 26369729 25244985 23537049 25348373 20798317 24495485 20075255 18362917 19820115 20353571 24845553 29403074 20566863 28004739 15632085 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26227816
EMBL
BABH01008405
ODYU01000461
SOQ35298.1
NWSH01004561
NWSH01003746
PCG64928.1
+ More
PCG65860.1 GDQN01009402 GDQN01008468 GDQN01007158 GDQN01003427 GDQN01003116 JAT81652.1 JAT82586.1 JAT83896.1 JAT87627.1 JAT87938.1 KQ459984 KPJ19087.1 KX778609 AON96578.1 KQ458793 KPJ05008.1 RSAL01000037 RVE51237.1 AGBW02008897 OWR52245.1 GFDL01014491 JAV20554.1 DS231816 EDS37568.1 AJVK01010584 AJWK01030992 KQ414727 KOC62481.1 CH477890 EAT35303.1 JXUM01045473 KQ561440 KXJ78581.1 GFDF01008395 JAV05689.1 GGFJ01009022 MBW58163.1 GGFJ01009021 MBW58162.1 KK107119 QOIP01000009 EZA58670.1 RLU18868.1 KQ759851 OAD62557.1 ADMH02002138 ETN58334.1 KQ435821 KOX72421.1 KQ435127 KZC14810.1 GL888679 EGI58535.1 KQ981993 KYN31107.1 GL763054 EFZ20235.1 KQ976772 KYN08454.1 ADTU01027553 AAAB01008980 EAA13884.4 APCN01005756 CAQQ02144572 AXCN02000652 KZ288215 PBC32542.1 GDAI01001889 JAI15714.1 UFQS01000289 UFQS01001336 UFQT01000289 UFQT01001336 SSX02482.1 SSX10393.1 SSX22856.1 AXCM01001829 KQ982974 KYQ48582.1 KB632191 ERL89813.1 GDHF01013431 JAI38883.1 GAKP01004759 JAC54193.1 NEVH01000277 PNF43684.1 GL443520 EFN61963.1 GAMC01003464 JAC03092.1 GDHF01020861 JAI31453.1 KQ971372 EFA10451.1 GBYB01011382 GBYB01011383 JAG81149.1 JAG81150.1 GECU01020775 JAS86931.1 GEBQ01006543 JAT33434.1 GECZ01012398 JAS57371.1 EZ423945 ADD20221.1 KK853235 KDR09543.1 PYGN01001870 PSN32407.1 APGK01050704 KB741175 ENN73214.1 JH432011 GDRN01094117 JAI59759.1 GDRN01094116 JAI59760.1 DS235813 EEB17451.1 EZ423946 ADD20222.1 GEDC01014913 JAS22385.1 GFTR01003645 JAW12781.1 GEZM01048958 JAV76244.1 CH379062 EAL32836.2 CH479184 EDW37315.1 CH902620 EDV31603.1 CM000361 CM002910 EDX04034.1 KMY88658.1 GGFM01005589 MBW26340.1 CCAG010007618 AE014134 AY061214 AAF52462.2 AAL28762.1 CH480818 EDW51934.1 CM000157 EDW87814.1 CH963920 EDW78008.2 CP012523 ALC38640.1 ACPB03021036 ACPB03021037 GAHY01001176 JAA76334.1 CH933807 EDW12563.1 CH916368 EDW03671.1 CH940649 EDW64638.1 JTDY01001772 KOB72926.1
PCG65860.1 GDQN01009402 GDQN01008468 GDQN01007158 GDQN01003427 GDQN01003116 JAT81652.1 JAT82586.1 JAT83896.1 JAT87627.1 JAT87938.1 KQ459984 KPJ19087.1 KX778609 AON96578.1 KQ458793 KPJ05008.1 RSAL01000037 RVE51237.1 AGBW02008897 OWR52245.1 GFDL01014491 JAV20554.1 DS231816 EDS37568.1 AJVK01010584 AJWK01030992 KQ414727 KOC62481.1 CH477890 EAT35303.1 JXUM01045473 KQ561440 KXJ78581.1 GFDF01008395 JAV05689.1 GGFJ01009022 MBW58163.1 GGFJ01009021 MBW58162.1 KK107119 QOIP01000009 EZA58670.1 RLU18868.1 KQ759851 OAD62557.1 ADMH02002138 ETN58334.1 KQ435821 KOX72421.1 KQ435127 KZC14810.1 GL888679 EGI58535.1 KQ981993 KYN31107.1 GL763054 EFZ20235.1 KQ976772 KYN08454.1 ADTU01027553 AAAB01008980 EAA13884.4 APCN01005756 CAQQ02144572 AXCN02000652 KZ288215 PBC32542.1 GDAI01001889 JAI15714.1 UFQS01000289 UFQS01001336 UFQT01000289 UFQT01001336 SSX02482.1 SSX10393.1 SSX22856.1 AXCM01001829 KQ982974 KYQ48582.1 KB632191 ERL89813.1 GDHF01013431 JAI38883.1 GAKP01004759 JAC54193.1 NEVH01000277 PNF43684.1 GL443520 EFN61963.1 GAMC01003464 JAC03092.1 GDHF01020861 JAI31453.1 KQ971372 EFA10451.1 GBYB01011382 GBYB01011383 JAG81149.1 JAG81150.1 GECU01020775 JAS86931.1 GEBQ01006543 JAT33434.1 GECZ01012398 JAS57371.1 EZ423945 ADD20221.1 KK853235 KDR09543.1 PYGN01001870 PSN32407.1 APGK01050704 KB741175 ENN73214.1 JH432011 GDRN01094117 JAI59759.1 GDRN01094116 JAI59760.1 DS235813 EEB17451.1 EZ423946 ADD20222.1 GEDC01014913 JAS22385.1 GFTR01003645 JAW12781.1 GEZM01048958 JAV76244.1 CH379062 EAL32836.2 CH479184 EDW37315.1 CH902620 EDV31603.1 CM000361 CM002910 EDX04034.1 KMY88658.1 GGFM01005589 MBW26340.1 CCAG010007618 AE014134 AY061214 AAF52462.2 AAL28762.1 CH480818 EDW51934.1 CM000157 EDW87814.1 CH963920 EDW78008.2 CP012523 ALC38640.1 ACPB03021036 ACPB03021037 GAHY01001176 JAA76334.1 CH933807 EDW12563.1 CH916368 EDW03671.1 CH940649 EDW64638.1 JTDY01001772 KOB72926.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000002320 UP000075880 UP000092462 UP000092461 UP000053825 UP000008820 UP000069940 UP000249989 UP000005203 UP000075885 UP000095301 UP000053097 UP000279307 UP000095300 UP000069272 UP000000673 UP000053105 UP000076502 UP000007755 UP000078541 UP000078542 UP000005205 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000015102 UP000075886 UP000075903 UP000242457 UP000076408 UP000075883 UP000075809 UP000030742 UP000075920 UP000235965 UP000000311 UP000075881 UP000002358 UP000007266 UP000091820 UP000092443 UP000078200 UP000027135 UP000245037 UP000019118 UP000075900 UP000009046 UP000001819 UP000008744 UP000007801 UP000192221 UP000000304 UP000092444 UP000000803 UP000001292 UP000002282 UP000007798 UP000092553 UP000015103 UP000009192 UP000001070 UP000008792 UP000192223 UP000085678 UP000037510
UP000002320 UP000075880 UP000092462 UP000092461 UP000053825 UP000008820 UP000069940 UP000249989 UP000005203 UP000075885 UP000095301 UP000053097 UP000279307 UP000095300 UP000069272 UP000000673 UP000053105 UP000076502 UP000007755 UP000078541 UP000078542 UP000005205 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075884 UP000015102 UP000075886 UP000075903 UP000242457 UP000076408 UP000075883 UP000075809 UP000030742 UP000075920 UP000235965 UP000000311 UP000075881 UP000002358 UP000007266 UP000091820 UP000092443 UP000078200 UP000027135 UP000245037 UP000019118 UP000075900 UP000009046 UP000001819 UP000008744 UP000007801 UP000192221 UP000000304 UP000092444 UP000000803 UP000001292 UP000002282 UP000007798 UP000092553 UP000015103 UP000009192 UP000001070 UP000008792 UP000192223 UP000085678 UP000037510
Interpro
SUPFAM
SSF103506
SSF103506
Gene 3D
ProteinModelPortal
H9JB02
A0A2H1V376
A0A2A4J341
A0A1E1WAA2
A0A194RNK2
A0A1C9EGF5
+ More
A0A194QHI5 A0A3S2NX48 A0A212FEQ9 A0A1Q3EZ26 B0VZZ6 A0A182JFM0 A0A1B0D2C8 A0A1B0CVQ1 A0A0L7QVG5 Q16LU6 A0A182G6R9 A0A1L8DGV8 A0A087ZWB7 A0A182PKV2 A0A2M4BYQ4 A0A1I8NA85 A0A2M4BYS3 A0A026WRK6 A0A1I8NRD6 A0A310SS33 A0A182F4D3 W5J696 A0A0N0U4R3 A0A154PUD2 F4X4W2 A0A195ET69 E9IGW2 A0A151IQL1 A0A158NVW3 A0A182TYY1 A0A182WZD9 A0A182L669 Q7Q1S4 A0A182ICS8 A0A182NAX8 T1GFC6 A0A182Q6F0 A0A182VD91 A0A2A3EMP5 A0A0K8TMY6 A0A182Y8B3 A0A336L1Y9 A0A182M7K7 A0A151WL25 U4UBW2 A0A182VTW6 A0A0K8VJ50 A0A034WKC8 A0A2J7RS70 E2AX80 W8C7R2 A0A182JNR6 A0A0K8UY36 K7IWJ2 D6WZM9 A0A0C9QAC1 A0A1A9WEA3 A0A1B6IJ38 A0A1B6MC32 A0A1B6G4R2 D3TR93 A0A1A9Y1I4 A0A1A9VYS8 A0A067QXQ0 A0A2P8XK76 N6SZK2 T1JBJ9 A0A182RGQ3 A0A0P4VV85 A0A0N7ZAT7 E0VVP5 D3TR94 A0A1B6D9X6 A0A224XXN6 A0A1Y1LRD1 Q29JX5 B4GKZ9 B3MKS2 A0A1W4UBD1 B4Q5A1 A0A2M3ZD09 A0A1B0FB53 Q9VM63 B4HY04 B4NZY7 B4N073 A0A0M4E7C1 R4G419 B4KJW0 B4JEF1 B4LRN6 A0A1W4XLN3 A0A1S3JLX6 A0A0L7LCJ9
A0A194QHI5 A0A3S2NX48 A0A212FEQ9 A0A1Q3EZ26 B0VZZ6 A0A182JFM0 A0A1B0D2C8 A0A1B0CVQ1 A0A0L7QVG5 Q16LU6 A0A182G6R9 A0A1L8DGV8 A0A087ZWB7 A0A182PKV2 A0A2M4BYQ4 A0A1I8NA85 A0A2M4BYS3 A0A026WRK6 A0A1I8NRD6 A0A310SS33 A0A182F4D3 W5J696 A0A0N0U4R3 A0A154PUD2 F4X4W2 A0A195ET69 E9IGW2 A0A151IQL1 A0A158NVW3 A0A182TYY1 A0A182WZD9 A0A182L669 Q7Q1S4 A0A182ICS8 A0A182NAX8 T1GFC6 A0A182Q6F0 A0A182VD91 A0A2A3EMP5 A0A0K8TMY6 A0A182Y8B3 A0A336L1Y9 A0A182M7K7 A0A151WL25 U4UBW2 A0A182VTW6 A0A0K8VJ50 A0A034WKC8 A0A2J7RS70 E2AX80 W8C7R2 A0A182JNR6 A0A0K8UY36 K7IWJ2 D6WZM9 A0A0C9QAC1 A0A1A9WEA3 A0A1B6IJ38 A0A1B6MC32 A0A1B6G4R2 D3TR93 A0A1A9Y1I4 A0A1A9VYS8 A0A067QXQ0 A0A2P8XK76 N6SZK2 T1JBJ9 A0A182RGQ3 A0A0P4VV85 A0A0N7ZAT7 E0VVP5 D3TR94 A0A1B6D9X6 A0A224XXN6 A0A1Y1LRD1 Q29JX5 B4GKZ9 B3MKS2 A0A1W4UBD1 B4Q5A1 A0A2M3ZD09 A0A1B0FB53 Q9VM63 B4HY04 B4NZY7 B4N073 A0A0M4E7C1 R4G419 B4KJW0 B4JEF1 B4LRN6 A0A1W4XLN3 A0A1S3JLX6 A0A0L7LCJ9
PDB
5VNB
E-value=2.95272e-53,
Score=524
Ontologies
KEGG
GO
Topology
Subcellular location
Nucleus
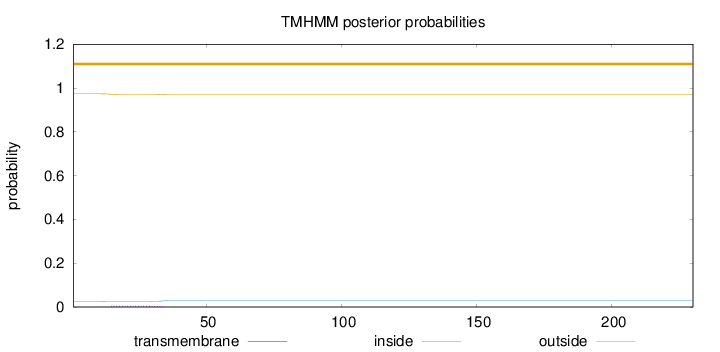
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09689
Exp number, first 60 AAs:
0.09689
Total prob of N-in:
0.02602
outside
1 - 230
Population Genetic Test Statistics
Pi
254.674887
Theta
198.647807
Tajima's D
0.867343
CLR
0
CSRT
0.622268886555672
Interpretation
Uncertain
