Gene
KWMTBOMO05437
Pre Gene Modal
BGIBMGA006690
Annotation
PREDICTED:_sin3_histone_deacetylase_corepressor_complex_component_SDS3_isoform_X2_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 4.745
Sequence
CDS
ATGCAACTGTTACGCGTCGCCTCGACTATGTCTTACCAAGGATCACCATATTCCGGCCCGGGGGATGAATACGATTTTGAAGACGATGGGTACGACGATTTAGACGACTTCAGAGACCGAGAAGAAACCGTCCCACCTCCTGCACCAGTGTTAGACAGCGACGAAGATACAGAAGAGGCAAGCGAGGCAGAACTACCGAACAACAATGAACCTCTGGAAGTTAAAGAACAAATATATCAAGATAAACTTGCCAATTTCAAGAAGCAACTGCAGCAATTGGAAGACAGCCTCCATCCTGAGTTCTTGCGTCGGGTGAAGAGGTTAGAACATCAGTTACAAGAGAGGTTACGATTGAACAGAATATACAAAGACCACATGTATGAAGTGGTAGACAGGGAGTATGTCGCCGAAAAAAAAGCTGCAGCTAAAGAATTTGAAGAGAAGAAGGTTGAGTTACGCGAGAACTTGCTCACCGATTTTGAAGACAAGAGGAAACTGATTGAGAGTGAACGACACAGCATGGAATTGAATGGTGATTCAATGGAGGTGAAACCGGTAATGAAGAGAATATTACGGAGGCGAGCCAACGAGCCGGCCCCGGCGCCGGAGAAGCGCCGCAAGCCGCTGGCCACCACGCTCACGTTCCAGCTGGACGAGCGCGACATCGAGGCCGACCTCCGCGCCATACAGCGGACCACCCCGCCCAGGCATCCCGTGCAGCACAACACCGCGCAGCCCAGGAAACATCTCAACTCTGGCAGTACATGCGACTCCCCTATTCGGGAGAGCGAGACCTGCGACACTCGTGTGGAAGACGGCAAGCTGTTGTACGAGAGGCGCTGGTACCACCGCGGGCAAAGCGTCTACGTCGAGGGACGGGACCTTCCCAAGTTCCCCGGACACATCCACGCCATTACAGATGAGGCGATATGGGTAAAGAAGACGAATGTGGAACGAATAAGGATATACATATCGCAGTTAGCGCGCGGTAAAGTGACGTTAAAACGACGAGCGTCGTAA
Protein
MQLLRVASTMSYQGSPYSGPGDEYDFEDDGYDDLDDFRDREETVPPPAPVLDSDEDTEEASEAELPNNNEPLEVKEQIYQDKLANFKKQLQQLEDSLHPEFLRRVKRLEHQLQERLRLNRIYKDHMYEVVDREYVAEKKAAAKEFEEKKVELRENLLTDFEDKRKLIESERHSMELNGDSMEVKPVMKRILRRRANEPAPAPEKRRKPLATTLTFQLDERDIEADLRAIQRTTPPRHPVQHNTAQPRKHLNSGSTCDSPIRESETCDTRVEDGKLLYERRWYHRGQSVYVEGRDLPKFPGHIHAITDEAIWVKKTNVERIRIYISQLARGKVTLKRRAS
Summary
Uniprot
H9JAZ5
A0A2A4JIY3
A0A2H1X2B3
A0A212F1N9
A0A194RMN8
A0A194QND9
+ More
A0A1C9EGH6 A0A1Y1L8Y1 A0A2J7PJV2 E2BWV9 A0A151XE53 A0A195DV81 D6X120 A0A087ZY99 A0A2A3EEK0 A0A067R4D4 A0A158NJV3 A0A195BJ01 A0A1W4W898 A0A151I810 A0A195F4M3 A0A026WPR2 A0A154PBD4 E1ZWX4 F4WIQ1 A0A0L7QSQ7 A0A0N0U601 A0A1A9ZBC8 D3TQ14 A0A1B0BPE2 A0A0C9QI57 Q17JC1 A0A1A9WB20 A0A0K8TKV2 A0A0L0BTB7 A0A1A9UHZ2 A0A0C9RM01 A0A1I8M6V3 N6TTU8 A0A1I8QDL0 A0A0A9XBZ9 A0A2R7WXH8 A0A336L954 A0A336MNU0 E0VKY1 Q29IV4 B4HAQ1 A0A1B0DR87 A0A069DSQ6 A0A224XFW2 B4R7S0 A0A0M4EJY1 A0A1L8DX93 A0A1L8DXC5 B4I776 B3NVQ1 A0A0A1XSE6 B4PZG2 Q9VWG2 B3MQA3 Q8MS11 B4MAS5 A0A034VGZ9 B4JME6 A0A0P4VXM2 R4G4R5 A0A0K8VEB5 W8BAJ2 A0A1W4VHE6 A0A1B6GLD0 A0A310SDE6 W8C1X1 A0A1B6I5G2 B4L810 B4NC23 A0A182I9K6 A0A293M384 A0A182LTK5 A0A1S3HIU2 A0A131XIL9 A0A224Z6M2 A0A1S3HK43 A0A131YA89 A0A2R5L6U3 A0A1A8B1Q6 A0A1A8MII7 A0A1A8IKM5 A0A1A8QMD8 A0A3Q3KXX4 A0A3Q0EG34 A0A023GIE6 G3MQ41 A0A023FLG0 A0A1E1XG82 A0A3P8QZJ0 I7GJR2 A0A1A8GIC7 A0A1A8U260 A0A1A8DCW4 A0A3Q2WP85
A0A1C9EGH6 A0A1Y1L8Y1 A0A2J7PJV2 E2BWV9 A0A151XE53 A0A195DV81 D6X120 A0A087ZY99 A0A2A3EEK0 A0A067R4D4 A0A158NJV3 A0A195BJ01 A0A1W4W898 A0A151I810 A0A195F4M3 A0A026WPR2 A0A154PBD4 E1ZWX4 F4WIQ1 A0A0L7QSQ7 A0A0N0U601 A0A1A9ZBC8 D3TQ14 A0A1B0BPE2 A0A0C9QI57 Q17JC1 A0A1A9WB20 A0A0K8TKV2 A0A0L0BTB7 A0A1A9UHZ2 A0A0C9RM01 A0A1I8M6V3 N6TTU8 A0A1I8QDL0 A0A0A9XBZ9 A0A2R7WXH8 A0A336L954 A0A336MNU0 E0VKY1 Q29IV4 B4HAQ1 A0A1B0DR87 A0A069DSQ6 A0A224XFW2 B4R7S0 A0A0M4EJY1 A0A1L8DX93 A0A1L8DXC5 B4I776 B3NVQ1 A0A0A1XSE6 B4PZG2 Q9VWG2 B3MQA3 Q8MS11 B4MAS5 A0A034VGZ9 B4JME6 A0A0P4VXM2 R4G4R5 A0A0K8VEB5 W8BAJ2 A0A1W4VHE6 A0A1B6GLD0 A0A310SDE6 W8C1X1 A0A1B6I5G2 B4L810 B4NC23 A0A182I9K6 A0A293M384 A0A182LTK5 A0A1S3HIU2 A0A131XIL9 A0A224Z6M2 A0A1S3HK43 A0A131YA89 A0A2R5L6U3 A0A1A8B1Q6 A0A1A8MII7 A0A1A8IKM5 A0A1A8QMD8 A0A3Q3KXX4 A0A3Q0EG34 A0A023GIE6 G3MQ41 A0A023FLG0 A0A1E1XG82 A0A3P8QZJ0 I7GJR2 A0A1A8GIC7 A0A1A8U260 A0A1A8DCW4 A0A3Q2WP85
Pubmed
19121390
22118469
26354079
28004739
20798317
18362917
+ More
19820115 24845553 21347285 24508170 30249741 21719571 20353571 17510324 26369729 26108605 25315136 23537049 25401762 26823975 20566863 15632085 17994087 26334808 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 27129103 24495485 28049606 28797301 22216098 28503490 17194215
19820115 24845553 21347285 24508170 30249741 21719571 20353571 17510324 26369729 26108605 25315136 23537049 25401762 26823975 20566863 15632085 17994087 26334808 25830018 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 27129103 24495485 28049606 28797301 22216098 28503490 17194215
EMBL
BABH01008382
BABH01008383
NWSH01001403
PCG71362.1
ODYU01012946
SOQ59500.1
+ More
AGBW02010841 OWR47652.1 KQ459984 KPJ19093.1 KQ458793 KPJ05001.1 KX778609 AON96585.1 GEZM01063114 JAV69298.1 NEVH01024944 PNF16613.1 GL451188 EFN79823.1 KQ982254 KYQ58600.1 KQ980304 KYN16756.1 KQ971367 EFA09398.1 KZ288266 PBC30140.1 KK852704 KDR18046.1 ADTU01018395 ADTU01018396 KQ976465 KYM84337.1 KQ978390 KYM94274.1 KQ981820 KYN35336.1 KK107135 QOIP01000006 EZA58047.1 RLU21364.1 KQ434868 KZC09219.1 GL434909 EFN74314.1 GL888176 EGI65987.1 KQ414756 KOC61665.1 KQ435760 KOX75602.1 CCAG010013939 EZ423516 ADD19792.1 JXJN01017891 GBYB01014403 JAG84170.1 CH477233 EAT46783.1 GDAI01002644 JAI14959.1 JRES01001383 KNC23233.1 GBYB01014402 JAG84169.1 APGK01019138 KB740092 KB632429 ENN81468.1 ERL95425.1 GBHO01026453 GBRD01007184 GDHC01005911 JAG17151.1 JAG58637.1 JAQ12718.1 KK855781 PTY23870.1 UFQS01002611 UFQT01002611 SSX14356.1 SSX33767.1 UFQT01001300 SSX29897.1 DS235255 EEB14037.1 CH379063 EAL32549.1 CH479242 EDW37662.1 AJVK01008968 GBGD01002182 JAC86707.1 GFTR01005060 JAW11366.1 CM000366 EDX18403.1 CP012528 ALC49554.1 GFDF01003072 JAV11012.1 GFDF01003179 JAV10905.1 CH480823 EDW56174.1 CH954180 EDV46716.1 GBXI01000819 JAD13473.1 CM000162 EDX02118.1 AE014298 BT044333 AAF48979.1 ACH92398.1 CH902621 EDV44529.1 AY119157 AAM51017.1 CH940655 EDW66334.2 GAKP01017565 JAC41387.1 CH916371 EDV92471.1 GDKW01001184 JAI55411.1 ACPB03000489 GAHY01000551 JAA76959.1 GDHF01015045 JAI37269.1 GAMC01010853 GAMC01010849 JAB95702.1 GECZ01006520 GECZ01004035 JAS63249.1 JAS65734.1 KQ773953 OAD52339.1 GAMC01010851 GAMC01010847 JAB95704.1 GECU01025537 JAS82169.1 CH933814 EDW05585.1 CH964239 EDW82382.1 APCN01000989 GFWV01009822 MAA34551.1 AXCM01001185 GEFH01002516 JAP66065.1 GFPF01011775 MAA22921.1 GEFM01000338 JAP75458.1 GGLE01001100 MBY05226.1 HADY01022714 SBP61199.1 HAEF01015035 SBR56194.1 HAED01011402 SBQ97721.1 HAEH01012233 SBR94279.1 GBBM01001744 JAC33674.1 JO843992 AEO35609.1 GBBK01002979 JAC21503.1 GFAC01001082 JAT98106.1 AB170399 BAE87462.1 HAEB01013370 HAEC01003468 SBQ71545.1 HAEJ01001703 SBS42160.1 HADZ01017799 HAEA01003452 SBQ31932.1
AGBW02010841 OWR47652.1 KQ459984 KPJ19093.1 KQ458793 KPJ05001.1 KX778609 AON96585.1 GEZM01063114 JAV69298.1 NEVH01024944 PNF16613.1 GL451188 EFN79823.1 KQ982254 KYQ58600.1 KQ980304 KYN16756.1 KQ971367 EFA09398.1 KZ288266 PBC30140.1 KK852704 KDR18046.1 ADTU01018395 ADTU01018396 KQ976465 KYM84337.1 KQ978390 KYM94274.1 KQ981820 KYN35336.1 KK107135 QOIP01000006 EZA58047.1 RLU21364.1 KQ434868 KZC09219.1 GL434909 EFN74314.1 GL888176 EGI65987.1 KQ414756 KOC61665.1 KQ435760 KOX75602.1 CCAG010013939 EZ423516 ADD19792.1 JXJN01017891 GBYB01014403 JAG84170.1 CH477233 EAT46783.1 GDAI01002644 JAI14959.1 JRES01001383 KNC23233.1 GBYB01014402 JAG84169.1 APGK01019138 KB740092 KB632429 ENN81468.1 ERL95425.1 GBHO01026453 GBRD01007184 GDHC01005911 JAG17151.1 JAG58637.1 JAQ12718.1 KK855781 PTY23870.1 UFQS01002611 UFQT01002611 SSX14356.1 SSX33767.1 UFQT01001300 SSX29897.1 DS235255 EEB14037.1 CH379063 EAL32549.1 CH479242 EDW37662.1 AJVK01008968 GBGD01002182 JAC86707.1 GFTR01005060 JAW11366.1 CM000366 EDX18403.1 CP012528 ALC49554.1 GFDF01003072 JAV11012.1 GFDF01003179 JAV10905.1 CH480823 EDW56174.1 CH954180 EDV46716.1 GBXI01000819 JAD13473.1 CM000162 EDX02118.1 AE014298 BT044333 AAF48979.1 ACH92398.1 CH902621 EDV44529.1 AY119157 AAM51017.1 CH940655 EDW66334.2 GAKP01017565 JAC41387.1 CH916371 EDV92471.1 GDKW01001184 JAI55411.1 ACPB03000489 GAHY01000551 JAA76959.1 GDHF01015045 JAI37269.1 GAMC01010853 GAMC01010849 JAB95702.1 GECZ01006520 GECZ01004035 JAS63249.1 JAS65734.1 KQ773953 OAD52339.1 GAMC01010851 GAMC01010847 JAB95704.1 GECU01025537 JAS82169.1 CH933814 EDW05585.1 CH964239 EDW82382.1 APCN01000989 GFWV01009822 MAA34551.1 AXCM01001185 GEFH01002516 JAP66065.1 GFPF01011775 MAA22921.1 GEFM01000338 JAP75458.1 GGLE01001100 MBY05226.1 HADY01022714 SBP61199.1 HAEF01015035 SBR56194.1 HAED01011402 SBQ97721.1 HAEH01012233 SBR94279.1 GBBM01001744 JAC33674.1 JO843992 AEO35609.1 GBBK01002979 JAC21503.1 GFAC01001082 JAT98106.1 AB170399 BAE87462.1 HAEB01013370 HAEC01003468 SBQ71545.1 HAEJ01001703 SBS42160.1 HADZ01017799 HAEA01003452 SBQ31932.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000008237 UP000075809 UP000078492 UP000007266 UP000005203 UP000242457 UP000027135 UP000005205 UP000078540 UP000192223 UP000078542 UP000078541 UP000053097 UP000279307 UP000076502 UP000000311 UP000007755 UP000053825 UP000053105 UP000092445 UP000092444 UP000092460 UP000008820 UP000091820 UP000037069 UP000078200 UP000095301 UP000019118 UP000030742 UP000095300 UP000009046 UP000001819 UP000008744 UP000092462 UP000000304 UP000092553 UP000001292 UP000008711 UP000002282 UP000000803 UP000007801 UP000008792 UP000001070 UP000015103 UP000192221 UP000009192 UP000007798 UP000075840 UP000075883 UP000085678 UP000261640 UP000189704 UP000265100 UP000264840
UP000008237 UP000075809 UP000078492 UP000007266 UP000005203 UP000242457 UP000027135 UP000005205 UP000078540 UP000192223 UP000078542 UP000078541 UP000053097 UP000279307 UP000076502 UP000000311 UP000007755 UP000053825 UP000053105 UP000092445 UP000092444 UP000092460 UP000008820 UP000091820 UP000037069 UP000078200 UP000095301 UP000019118 UP000030742 UP000095300 UP000009046 UP000001819 UP000008744 UP000092462 UP000000304 UP000092553 UP000001292 UP000008711 UP000002282 UP000000803 UP000007801 UP000008792 UP000001070 UP000015103 UP000192221 UP000009192 UP000007798 UP000075840 UP000075883 UP000085678 UP000261640 UP000189704 UP000265100 UP000264840
Gene 3D
ProteinModelPortal
H9JAZ5
A0A2A4JIY3
A0A2H1X2B3
A0A212F1N9
A0A194RMN8
A0A194QND9
+ More
A0A1C9EGH6 A0A1Y1L8Y1 A0A2J7PJV2 E2BWV9 A0A151XE53 A0A195DV81 D6X120 A0A087ZY99 A0A2A3EEK0 A0A067R4D4 A0A158NJV3 A0A195BJ01 A0A1W4W898 A0A151I810 A0A195F4M3 A0A026WPR2 A0A154PBD4 E1ZWX4 F4WIQ1 A0A0L7QSQ7 A0A0N0U601 A0A1A9ZBC8 D3TQ14 A0A1B0BPE2 A0A0C9QI57 Q17JC1 A0A1A9WB20 A0A0K8TKV2 A0A0L0BTB7 A0A1A9UHZ2 A0A0C9RM01 A0A1I8M6V3 N6TTU8 A0A1I8QDL0 A0A0A9XBZ9 A0A2R7WXH8 A0A336L954 A0A336MNU0 E0VKY1 Q29IV4 B4HAQ1 A0A1B0DR87 A0A069DSQ6 A0A224XFW2 B4R7S0 A0A0M4EJY1 A0A1L8DX93 A0A1L8DXC5 B4I776 B3NVQ1 A0A0A1XSE6 B4PZG2 Q9VWG2 B3MQA3 Q8MS11 B4MAS5 A0A034VGZ9 B4JME6 A0A0P4VXM2 R4G4R5 A0A0K8VEB5 W8BAJ2 A0A1W4VHE6 A0A1B6GLD0 A0A310SDE6 W8C1X1 A0A1B6I5G2 B4L810 B4NC23 A0A182I9K6 A0A293M384 A0A182LTK5 A0A1S3HIU2 A0A131XIL9 A0A224Z6M2 A0A1S3HK43 A0A131YA89 A0A2R5L6U3 A0A1A8B1Q6 A0A1A8MII7 A0A1A8IKM5 A0A1A8QMD8 A0A3Q3KXX4 A0A3Q0EG34 A0A023GIE6 G3MQ41 A0A023FLG0 A0A1E1XG82 A0A3P8QZJ0 I7GJR2 A0A1A8GIC7 A0A1A8U260 A0A1A8DCW4 A0A3Q2WP85
A0A1C9EGH6 A0A1Y1L8Y1 A0A2J7PJV2 E2BWV9 A0A151XE53 A0A195DV81 D6X120 A0A087ZY99 A0A2A3EEK0 A0A067R4D4 A0A158NJV3 A0A195BJ01 A0A1W4W898 A0A151I810 A0A195F4M3 A0A026WPR2 A0A154PBD4 E1ZWX4 F4WIQ1 A0A0L7QSQ7 A0A0N0U601 A0A1A9ZBC8 D3TQ14 A0A1B0BPE2 A0A0C9QI57 Q17JC1 A0A1A9WB20 A0A0K8TKV2 A0A0L0BTB7 A0A1A9UHZ2 A0A0C9RM01 A0A1I8M6V3 N6TTU8 A0A1I8QDL0 A0A0A9XBZ9 A0A2R7WXH8 A0A336L954 A0A336MNU0 E0VKY1 Q29IV4 B4HAQ1 A0A1B0DR87 A0A069DSQ6 A0A224XFW2 B4R7S0 A0A0M4EJY1 A0A1L8DX93 A0A1L8DXC5 B4I776 B3NVQ1 A0A0A1XSE6 B4PZG2 Q9VWG2 B3MQA3 Q8MS11 B4MAS5 A0A034VGZ9 B4JME6 A0A0P4VXM2 R4G4R5 A0A0K8VEB5 W8BAJ2 A0A1W4VHE6 A0A1B6GLD0 A0A310SDE6 W8C1X1 A0A1B6I5G2 B4L810 B4NC23 A0A182I9K6 A0A293M384 A0A182LTK5 A0A1S3HIU2 A0A131XIL9 A0A224Z6M2 A0A1S3HK43 A0A131YA89 A0A2R5L6U3 A0A1A8B1Q6 A0A1A8MII7 A0A1A8IKM5 A0A1A8QMD8 A0A3Q3KXX4 A0A3Q0EG34 A0A023GIE6 G3MQ41 A0A023FLG0 A0A1E1XG82 A0A3P8QZJ0 I7GJR2 A0A1A8GIC7 A0A1A8U260 A0A1A8DCW4 A0A3Q2WP85
Ontologies
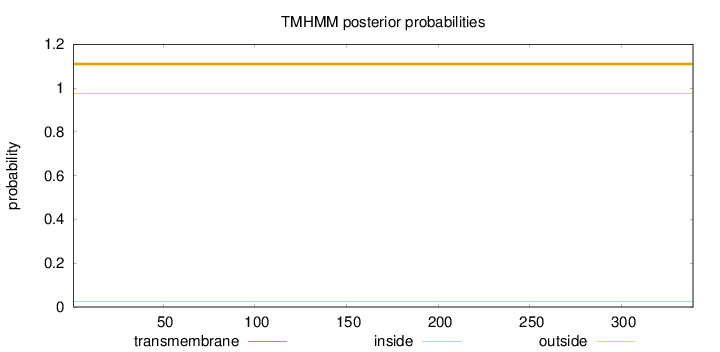
Topology
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02370
outside
1 - 339
Population Genetic Test Statistics
Pi
272.184905
Theta
206.163869
Tajima's D
0.922086
CLR
0.004022
CSRT
0.64176791160442
Interpretation
Uncertain
