Pre Gene Modal
BGIBMGA006649
Annotation
Uncharacterized_protein_OBRU01_02299_[Operophtera_brumata]
Full name
Protein CLP1 homolog
Location in the cell
Nuclear Reliability : 2.236
Sequence
CDS
ATGTCGTCCGAAATGTTAATACGAGAGCCACCGCCCACAAGAATGATTTTCAAGGCAATCGACGAACAAAGTTCTGAGGAGCCTCCGATGGAGGAATGGTCTTCGTGTCAACTGGAAGTCAATGAAACTAATGTGAAGAAAGCCTTGTTTCAAGAGGGTCTTTATGCACCAGGAAGTGGAGAAATATGCACTAAATACATTACTGTTTCTGACAGCTCAGTTCTAAGACACCCATATTTTAACTACCCTGGAGTCATCGATCCTGGTATTGAAAAAGCTTTGCTACAACCAGAACCTCCTATAATATACCCTGACGATGGTCAGGTATTATATCTTGAGCTGTGCAAAGAAATGGGACAGTGCCCAGTTAGAATGTTCCATAAAGGACTACTGAACGATGTGATAGATTTGAGGTATTACGGTGTAAATCCAAACAGTGTCCGATGCATGGGCATTGCAATGAAATTTAATAATTTTGTCAAAGTTTTAAATTTAACCGATAATTTCTTGAATAACGACGCTTGTTACCACCTTGGTGAGATGCTTACTACGAATTCGTGTTTACGAGAATTGAATCTTTGCGGATGTCGCATTGGTGTTGAAGGTCTTCGAAGATTACTTCCCGGCCTCATCGTAAATAGATCTTTGAAAGTATTGAATCTTAATAAAAATCAGCTGGGTGATACTGGGATACAATTAGTAGCAAGAGGTATAGTTCGCGGTATGGACGTCGAAAATTTATATCTTAGTTATAATGATCTCAGTGCTAATGCAGCCAATATATTAACTGAAGCTTTCAATAGGCACAACAAACTTGTTTTACTAGATCTGTCTTGGAATAATCTGTACTCGCCTGCAGAGGTATACGTTTTATTAACTAAACTTGCTCAAAATGAATTTTTGCAGACATTAGATTTATCTTGGAATTCTTTAGCCGGAGCCAGGATCGGTAATGCTATAAAAATATTAATGAAAGCTCCTAATTTGCAAAATTTGAACTTAAGCAACAATAGATTAACCGGTGATGCTATATCCAACCTGCTGAAGAATATAGAAAAACCCCAGAATCTCGTTCTGAATCTCTCGTACAATCCTATGACACCAACTGATGCCTTATCCATCCTTAATAAGTTGAAGTTACCTGAAGTGAAACTAAATACACTTTTGTTAGACAACGTTTTAGTGGGGTGCGAATTTTTGGAGGCTCTTAAAGAAATCAGAGAACTTGAATTTCGTAGGGATACAATCATAACCCACGGAGGATTAACTAATGGCAACAAGCCTACTGGACCTGATCCTCGTGATGTGGTGTTAAACAGAGCTGATTTTCTGTGCCTGAATCCAAAGAAACAACCTATAGACATTGCTTTAGTATTATTGAAGCTACTAAAAACAGGTAACAAGATTATGAGTATAAAGGACTTTGCGAAAACTCTTCGAAGGTCCGGTGTTCCTCTGGATCCAGATTTAATTGAAGAAATGACAGATGCGTTCGCAGGACCACGGACAACTAAAGCTAAAGTAATCGATCTAAAGCTTGTCGCAGATTACATTAAAAGGAAATGGCCTGAGAGAAAATTGCCCGTTACGCCTCCACCTGAACCAATACGAAAGAAGAGCAAAAGGAAGGCGAAAAAGAAACATTGA
Protein
MSSEMLIREPPPTRMIFKAIDEQSSEEPPMEEWSSCQLEVNETNVKKALFQEGLYAPGSGEICTKYITVSDSSVLRHPYFNYPGVIDPGIEKALLQPEPPIIYPDDGQVLYLELCKEMGQCPVRMFHKGLLNDVIDLRYYGVNPNSVRCMGIAMKFNNFVKVLNLTDNFLNNDACYHLGEMLTTNSCLRELNLCGCRIGVEGLRRLLPGLIVNRSLKVLNLNKNQLGDTGIQLVARGIVRGMDVENLYLSYNDLSANAANILTEAFNRHNKLVLLDLSWNNLYSPAEVYVLLTKLAQNEFLQTLDLSWNSLAGARIGNAIKILMKAPNLQNLNLSNNRLTGDAISNLLKNIEKPQNLVLNLSYNPMTPTDALSILNKLKLPEVKLNTLLLDNVLVGCEFLEALKEIRELEFRRDTIITHGGLTNGNKPTGPDPRDVVLNRADFLCLNPKKQPIDIALVLLKLLKTGNKIMSIKDFAKTLRRSGVPLDPDLIEEMTDAFAGPRTTKAKVIDLKLVADYIKRKWPERKLPVTPPPEPIRKKSKRKAKKKH
Summary
Description
Required for endonucleolytic cleavage during polyadenylation-dependent pre-mRNA 3'-end formation.
Similarity
Belongs to the Clp1 family. Clp1 subfamily.
Uniprot
H9JAV4
A0A0L7LTB1
A0A1C9EGH4
A0A0L7LSM5
A0A2H1WGQ1
A0A212F1N8
+ More
A0A194QHL3 A0A194RNL2 A0A194QJ93 A0A194QND4 A0A2H1W1H8 A0A194RMP2 H9JB25 H9JN06 A0A212FAK7 A0A2H1WXC4 E2BHS2 A0A1B6ETI4 A0A0L7QMC8 A0A0M9A4E4 A0A195CMP5 A0A088AB53 A0A151XCR4 A0A151JC12 A0A2A3EPX5 A0A026W7F1 A0A151I302 A0A1Y1L9F8 A0A1B6KJN2 A0A195F9T4 N6SZA8 U4URG0 A0A232EGL8 F4WYU4 A0A139WEM6 A0A2B4SG47 H3B3F3 M3XJM7 A0A3N0YNI6 F1QNH6 U3J029 A0A210PQF7 A0A384AB95 A0A0L8GFQ3 A0A1W4ZLD1 A0A0L8GFJ2 A0A2U3VVE7 A0A1L8I070 C3Z2D7 A0A2Y9NQD9 A0A151MLE8 A0A1S3J7G8 A0A1S3K5K9 A0A060XL28 A0A2Y9DAP5 A0A3P8XQC9 A0A1L8F9C2 A0A2Y9JJR4 A0A341ASK2 A0A3B4TGU6 W5NW24 W5L4F3 L8IZK9 A0A226NXL8 A0A2U9CJ39 A0A3P9Q2E3 A0A2Y9ERU1 A0A1S3HY18 A0A087Y8N0 A0A384D334 A0A3Q7NYF7 H3BA37 M3YC36 A0A2I4CZ82 A0A2K6SSK3 W4XPE5 A0A2Y9GBX7 A0A2K5DXY4 A0A1U8CZ56 A0A3Q1ETB7 A0A3Q7Y8Z0 S9WSB8
A0A194QHL3 A0A194RNL2 A0A194QJ93 A0A194QND4 A0A2H1W1H8 A0A194RMP2 H9JB25 H9JN06 A0A212FAK7 A0A2H1WXC4 E2BHS2 A0A1B6ETI4 A0A0L7QMC8 A0A0M9A4E4 A0A195CMP5 A0A088AB53 A0A151XCR4 A0A151JC12 A0A2A3EPX5 A0A026W7F1 A0A151I302 A0A1Y1L9F8 A0A1B6KJN2 A0A195F9T4 N6SZA8 U4URG0 A0A232EGL8 F4WYU4 A0A139WEM6 A0A2B4SG47 H3B3F3 M3XJM7 A0A3N0YNI6 F1QNH6 U3J029 A0A210PQF7 A0A384AB95 A0A0L8GFQ3 A0A1W4ZLD1 A0A0L8GFJ2 A0A2U3VVE7 A0A1L8I070 C3Z2D7 A0A2Y9NQD9 A0A151MLE8 A0A1S3J7G8 A0A1S3K5K9 A0A060XL28 A0A2Y9DAP5 A0A3P8XQC9 A0A1L8F9C2 A0A2Y9JJR4 A0A341ASK2 A0A3B4TGU6 W5NW24 W5L4F3 L8IZK9 A0A226NXL8 A0A2U9CJ39 A0A3P9Q2E3 A0A2Y9ERU1 A0A1S3HY18 A0A087Y8N0 A0A384D334 A0A3Q7NYF7 H3BA37 M3YC36 A0A2I4CZ82 A0A2K6SSK3 W4XPE5 A0A2Y9GBX7 A0A2K5DXY4 A0A1U8CZ56 A0A3Q1ETB7 A0A3Q7Y8Z0 S9WSB8
Pubmed
EMBL
BABH01008368
JTDY01000167
KOB78481.1
KX778609
AON96589.1
KOB78478.1
+ More
ODYU01008523 SOQ52197.1 AGBW02010841 OWR47646.1 KQ458793 KPJ04997.1 KQ459984 KPJ19097.1 KPJ04995.1 KPJ04996.1 ODYU01005735 SOQ46887.1 KPJ19098.1 BABH01008501 BABH01041387 AGBW02009461 OWR50759.1 ODYU01011363 SOQ57064.1 GL448324 EFN84755.1 GECZ01028464 JAS41305.1 KQ414894 KOC59686.1 KQ435736 KOX77005.1 KQ977565 KYN01950.1 KQ982298 KYQ58153.1 KQ979120 KYN22568.1 KZ288194 PBC33815.1 KK107429 EZA50959.1 KQ976508 KYM82782.1 GEZM01066689 JAV67727.1 GEBQ01028314 JAT11663.1 KQ981727 KYN37193.1 APGK01051086 APGK01051087 KB741181 ENN73089.1 KB632333 ERL92741.1 NNAY01004742 OXU17483.1 GL888463 EGI60598.1 KQ971354 KYB26428.1 LSMT01000062 PFX29674.1 AFYH01003060 AFYH01003061 RJVU01035218 ROL47769.1 AL935141 ADON01035317 NEDP02005558 OWF38686.1 KQ422015 KOF75783.1 KOF75781.1 CM004466 OCU01757.1 GG666574 EEN53188.1 AKHW03005800 KYO25378.1 FR905523 CDQ79942.1 CM004480 OCT68176.1 AMGL01104598 AMGL01104599 AMGL01104600 JH880545 ELR60794.1 AWGT02000321 OXB72027.1 CP026259 AWP16605.1 AYCK01003420 AFYH01024312 AEYP01004660 AAGJ04080353 KB017008 EPY81332.1
ODYU01008523 SOQ52197.1 AGBW02010841 OWR47646.1 KQ458793 KPJ04997.1 KQ459984 KPJ19097.1 KPJ04995.1 KPJ04996.1 ODYU01005735 SOQ46887.1 KPJ19098.1 BABH01008501 BABH01041387 AGBW02009461 OWR50759.1 ODYU01011363 SOQ57064.1 GL448324 EFN84755.1 GECZ01028464 JAS41305.1 KQ414894 KOC59686.1 KQ435736 KOX77005.1 KQ977565 KYN01950.1 KQ982298 KYQ58153.1 KQ979120 KYN22568.1 KZ288194 PBC33815.1 KK107429 EZA50959.1 KQ976508 KYM82782.1 GEZM01066689 JAV67727.1 GEBQ01028314 JAT11663.1 KQ981727 KYN37193.1 APGK01051086 APGK01051087 KB741181 ENN73089.1 KB632333 ERL92741.1 NNAY01004742 OXU17483.1 GL888463 EGI60598.1 KQ971354 KYB26428.1 LSMT01000062 PFX29674.1 AFYH01003060 AFYH01003061 RJVU01035218 ROL47769.1 AL935141 ADON01035317 NEDP02005558 OWF38686.1 KQ422015 KOF75783.1 KOF75781.1 CM004466 OCU01757.1 GG666574 EEN53188.1 AKHW03005800 KYO25378.1 FR905523 CDQ79942.1 CM004480 OCT68176.1 AMGL01104598 AMGL01104599 AMGL01104600 JH880545 ELR60794.1 AWGT02000321 OXB72027.1 CP026259 AWP16605.1 AYCK01003420 AFYH01024312 AEYP01004660 AAGJ04080353 KB017008 EPY81332.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053268
UP000053240
UP000008237
+ More
UP000053825 UP000053105 UP000078542 UP000005203 UP000075809 UP000078492 UP000242457 UP000053097 UP000078540 UP000078541 UP000019118 UP000030742 UP000215335 UP000007755 UP000007266 UP000225706 UP000008672 UP000000437 UP000016666 UP000242188 UP000261681 UP000053454 UP000192224 UP000245340 UP000186698 UP000001554 UP000248483 UP000050525 UP000085678 UP000193380 UP000248480 UP000265140 UP000248482 UP000252040 UP000261420 UP000002356 UP000018467 UP000198419 UP000246464 UP000242638 UP000028760 UP000261680 UP000286641 UP000000715 UP000192220 UP000233220 UP000007110 UP000248481 UP000233020 UP000189705 UP000257200 UP000286642
UP000053825 UP000053105 UP000078542 UP000005203 UP000075809 UP000078492 UP000242457 UP000053097 UP000078540 UP000078541 UP000019118 UP000030742 UP000215335 UP000007755 UP000007266 UP000225706 UP000008672 UP000000437 UP000016666 UP000242188 UP000261681 UP000053454 UP000192224 UP000245340 UP000186698 UP000001554 UP000248483 UP000050525 UP000085678 UP000193380 UP000248480 UP000265140 UP000248482 UP000252040 UP000261420 UP000002356 UP000018467 UP000198419 UP000246464 UP000242638 UP000028760 UP000261680 UP000286641 UP000000715 UP000192220 UP000233220 UP000007110 UP000248481 UP000233020 UP000189705 UP000257200 UP000286642
Pfam
Interpro
IPR032675
LRR_dom_sf
+ More
IPR001611 Leu-rich_rpt
IPR006553 Leu-rich_rpt_Cys-con_subtyp
IPR032324 Clp1_N
IPR027417 P-loop_NTPase
IPR038238 Clp1_C_sf
IPR038239 Clp1_N_sf
IPR032319 CLP1_P
IPR010655 Clp1_C
IPR028606 Clp1
IPR007526 SWIRM
IPR018247 EF_Hand_1_Ca_BS
IPR001005 SANT/Myb
IPR009057 Homeobox-like_sf
IPR017884 SANT_dom
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR003112 Olfac-like_dom
IPR001611 Leu-rich_rpt
IPR006553 Leu-rich_rpt_Cys-con_subtyp
IPR032324 Clp1_N
IPR027417 P-loop_NTPase
IPR038238 Clp1_C_sf
IPR038239 Clp1_N_sf
IPR032319 CLP1_P
IPR010655 Clp1_C
IPR028606 Clp1
IPR007526 SWIRM
IPR018247 EF_Hand_1_Ca_BS
IPR001005 SANT/Myb
IPR009057 Homeobox-like_sf
IPR017884 SANT_dom
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR003112 Olfac-like_dom
Gene 3D
ProteinModelPortal
H9JAV4
A0A0L7LTB1
A0A1C9EGH4
A0A0L7LSM5
A0A2H1WGQ1
A0A212F1N8
+ More
A0A194QHL3 A0A194RNL2 A0A194QJ93 A0A194QND4 A0A2H1W1H8 A0A194RMP2 H9JB25 H9JN06 A0A212FAK7 A0A2H1WXC4 E2BHS2 A0A1B6ETI4 A0A0L7QMC8 A0A0M9A4E4 A0A195CMP5 A0A088AB53 A0A151XCR4 A0A151JC12 A0A2A3EPX5 A0A026W7F1 A0A151I302 A0A1Y1L9F8 A0A1B6KJN2 A0A195F9T4 N6SZA8 U4URG0 A0A232EGL8 F4WYU4 A0A139WEM6 A0A2B4SG47 H3B3F3 M3XJM7 A0A3N0YNI6 F1QNH6 U3J029 A0A210PQF7 A0A384AB95 A0A0L8GFQ3 A0A1W4ZLD1 A0A0L8GFJ2 A0A2U3VVE7 A0A1L8I070 C3Z2D7 A0A2Y9NQD9 A0A151MLE8 A0A1S3J7G8 A0A1S3K5K9 A0A060XL28 A0A2Y9DAP5 A0A3P8XQC9 A0A1L8F9C2 A0A2Y9JJR4 A0A341ASK2 A0A3B4TGU6 W5NW24 W5L4F3 L8IZK9 A0A226NXL8 A0A2U9CJ39 A0A3P9Q2E3 A0A2Y9ERU1 A0A1S3HY18 A0A087Y8N0 A0A384D334 A0A3Q7NYF7 H3BA37 M3YC36 A0A2I4CZ82 A0A2K6SSK3 W4XPE5 A0A2Y9GBX7 A0A2K5DXY4 A0A1U8CZ56 A0A3Q1ETB7 A0A3Q7Y8Z0 S9WSB8
A0A194QHL3 A0A194RNL2 A0A194QJ93 A0A194QND4 A0A2H1W1H8 A0A194RMP2 H9JB25 H9JN06 A0A212FAK7 A0A2H1WXC4 E2BHS2 A0A1B6ETI4 A0A0L7QMC8 A0A0M9A4E4 A0A195CMP5 A0A088AB53 A0A151XCR4 A0A151JC12 A0A2A3EPX5 A0A026W7F1 A0A151I302 A0A1Y1L9F8 A0A1B6KJN2 A0A195F9T4 N6SZA8 U4URG0 A0A232EGL8 F4WYU4 A0A139WEM6 A0A2B4SG47 H3B3F3 M3XJM7 A0A3N0YNI6 F1QNH6 U3J029 A0A210PQF7 A0A384AB95 A0A0L8GFQ3 A0A1W4ZLD1 A0A0L8GFJ2 A0A2U3VVE7 A0A1L8I070 C3Z2D7 A0A2Y9NQD9 A0A151MLE8 A0A1S3J7G8 A0A1S3K5K9 A0A060XL28 A0A2Y9DAP5 A0A3P8XQC9 A0A1L8F9C2 A0A2Y9JJR4 A0A341ASK2 A0A3B4TGU6 W5NW24 W5L4F3 L8IZK9 A0A226NXL8 A0A2U9CJ39 A0A3P9Q2E3 A0A2Y9ERU1 A0A1S3HY18 A0A087Y8N0 A0A384D334 A0A3Q7NYF7 H3BA37 M3YC36 A0A2I4CZ82 A0A2K6SSK3 W4XPE5 A0A2Y9GBX7 A0A2K5DXY4 A0A1U8CZ56 A0A3Q1ETB7 A0A3Q7Y8Z0 S9WSB8
PDB
5IRL
E-value=2.83861e-08,
Score=141
Ontologies
GO
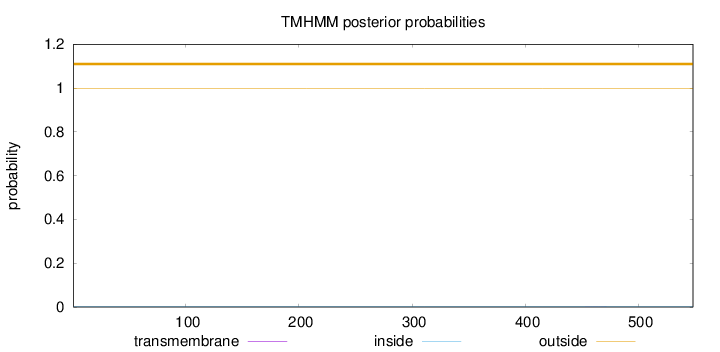
Topology
Subcellular location
Nucleus
Length:
548
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00794
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00188
outside
1 - 548
Population Genetic Test Statistics
Pi
132.751473
Theta
154.729598
Tajima's D
-0.455407
CLR
0.487768
CSRT
0.250087495625219
Interpretation
Uncertain
