Gene
KWMTBOMO05426 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006653
Annotation
PREDICTED:_39S_ribosomal_protein_L50?_mitochondrial_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 3.081
Sequence
CDS
ATGCTCAGAAATGTATTATCACCCTTACGTCGTGGAAGTTTCCAGGTCACATCTATCAGACAAGCTCAAAAAAAATATCCCAAAGTAGATAAAAAGGTTCAAGCAGCGGCAGAATCCTTAGCGGCAAGAGGCTTCCTACGTCCCACCAAGTCATGGGATCCACCTACAAACATCAATGAAACAATTCTGAAAATATGTGCAACATGTGGATTAAATGGAAATTCAGAATTTGAAGCATTGGACACAAAATTTACAGTTCTCAAAGCTTGTTATGAAGAAACAGGCCATAGTGTGCCTAATTCACTTTTGCATACAATTGAAACAGTTGATGACCTAACAGAATTTTACAAAACACCTGTGTACACAACTACACCGTTTGATGCTCTGAAGTTAATGGATCTACCCAAAAATCTACATGTTCAAAAAGATTATGTTAGATTTCATCCAGATACGGACACTTTATTCAATGGCGTCTCCGCTTTTCCGAAGAGTTCGACTATTGTAACCGGCCTTAAAGCACGGAAGAAGTATGAAGGCTACAATGCTAAAAGGAAATGGCCTTGA
Protein
MLRNVLSPLRRGSFQVTSIRQAQKKYPKVDKKVQAAAESLAARGFLRPTKSWDPPTNINETILKICATCGLNGNSEFEALDTKFTVLKACYEETGHSVPNSLLHTIETVDDLTEFYKTPVYTTTPFDALKLMDLPKNLHVQKDYVRFHPDTDTLFNGVSAFPKSSTIVTGLKARKKYEGYNAKRKWP
Summary
Uniprot
H9JAV8
A0A1E1WIF3
A0A2A4ITU9
A0A194RSQ6
A0A1C9EGH5
A0A2H1WGX9
+ More
A0A194QJ82 A0A212FFI3 A0A0L7LU49 A0A195F1C6 A0A158NFW3 A0A1A9ZMR6 A0A0L0CNR5 A0A1B0G6B8 A0A0M5J0W1 A0A0J7NX49 A0A1A9XTV2 A0A182VJA9 A0A182HM34 A0A182LJX1 A0A1A9UVU1 Q7Q760 A0A182V3N1 A0A151XC36 B4N3Q8 A0A182WQL9 Q9V3E3 A0A3B0J3J1 A0A034WGB2 B4HV34 A0A182MAR8 B3M4Y7 Q2M0G3 B4GRN5 B4QKB0 A0A0J9RRV2 A0A1L8EGQ1 U5ENH7 A0A3L8DLV2 A0A182IM66 A0A026WQT2 B4PK44 A0A1L8EH25 A0A023EKE2 A0A195E6R1 Q16IT0 W5JPY1 A0A2M3Z8P9 A0A2M3Z855 A0A1B0DDT5 A0A084W2X2 Q16I54 A0A0K8UUS4 A0A182Y0W2 A0A182T373 A0A1L8E0B4 A0A2M4BYQ9 A0A2M4BZV1 A0A2M4ARN5 A0A1Y1MDP8 A0A182LJL8 E2BDC1 A0A1W4UUR2 B3NFG5 T1DQN0 A0A2J7RJ36 A0A2M4ARM6 A0A2M4ARQ8 A0A336K691 A0A195CHC5 A0A2M4BYS4 A0A1A9W5R8 A0A1Q3F9P3 A0A2M4C0A4 B0WMS5 A0A1B0BBS8 A0A0K8TX66 E1ZYY3 B4LI70 B4KVR9 A0A0K8TQ72 A0A1W4W4B3 A0A1I8NXB2 A0A195B2M7 B4IY74 A0A2P8XYE1 A0A182KA93 A0A182PN78 A0A1I8N2J6 A0A1L8E1C9 E9J666 A0A182NM80 A0A0L7QJY9 A0A1B6BYY2 A0A067RTH4 A0A1B6M4T3 J9K2S1 A0A0P4VNV2 V9IFC4 T1I7I6
A0A194QJ82 A0A212FFI3 A0A0L7LU49 A0A195F1C6 A0A158NFW3 A0A1A9ZMR6 A0A0L0CNR5 A0A1B0G6B8 A0A0M5J0W1 A0A0J7NX49 A0A1A9XTV2 A0A182VJA9 A0A182HM34 A0A182LJX1 A0A1A9UVU1 Q7Q760 A0A182V3N1 A0A151XC36 B4N3Q8 A0A182WQL9 Q9V3E3 A0A3B0J3J1 A0A034WGB2 B4HV34 A0A182MAR8 B3M4Y7 Q2M0G3 B4GRN5 B4QKB0 A0A0J9RRV2 A0A1L8EGQ1 U5ENH7 A0A3L8DLV2 A0A182IM66 A0A026WQT2 B4PK44 A0A1L8EH25 A0A023EKE2 A0A195E6R1 Q16IT0 W5JPY1 A0A2M3Z8P9 A0A2M3Z855 A0A1B0DDT5 A0A084W2X2 Q16I54 A0A0K8UUS4 A0A182Y0W2 A0A182T373 A0A1L8E0B4 A0A2M4BYQ9 A0A2M4BZV1 A0A2M4ARN5 A0A1Y1MDP8 A0A182LJL8 E2BDC1 A0A1W4UUR2 B3NFG5 T1DQN0 A0A2J7RJ36 A0A2M4ARM6 A0A2M4ARQ8 A0A336K691 A0A195CHC5 A0A2M4BYS4 A0A1A9W5R8 A0A1Q3F9P3 A0A2M4C0A4 B0WMS5 A0A1B0BBS8 A0A0K8TX66 E1ZYY3 B4LI70 B4KVR9 A0A0K8TQ72 A0A1W4W4B3 A0A1I8NXB2 A0A195B2M7 B4IY74 A0A2P8XYE1 A0A182KA93 A0A182PN78 A0A1I8N2J6 A0A1L8E1C9 E9J666 A0A182NM80 A0A0L7QJY9 A0A1B6BYY2 A0A067RTH4 A0A1B6M4T3 J9K2S1 A0A0P4VNV2 V9IFC4 T1I7I6
Pubmed
19121390
26354079
22118469
26227816
21347285
26108605
+ More
20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 15632085 22936249 30249741 24508170 17550304 24945155 17510324 20920257 23761445 24438588 25244985 28004739 20798317 26369729 29403074 25315136 21282665 24845553 27129103
20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 15632085 22936249 30249741 24508170 17550304 24945155 17510324 20920257 23761445 24438588 25244985 28004739 20798317 26369729 29403074 25315136 21282665 24845553 27129103
EMBL
BABH01008349
GDQN01004280
JAT86774.1
NWSH01008413
PCG62563.1
KQ459984
+ More
KPJ19106.1 KX778609 AON96594.1 ODYU01008595 SOQ52321.1 KQ458793 KPJ04985.1 AGBW02008811 OWR52505.1 JTDY01000085 KOB78972.1 KQ981864 KYN34268.1 ADTU01014817 ADTU01014818 JRES01000234 KNC33094.1 CCAG010000599 CP012525 ALC44498.1 LBMM01001065 KMQ96990.1 APCN01004335 AAAB01008960 EAA11140.2 KQ982314 KYQ57924.1 CH964095 EDW79263.1 AF199369 AE014296 AY094750 AY113387 AAF09579.1 AAF50594.1 AAM11103.1 AAM29392.1 OUUW01000002 SPP76224.1 GAKP01004331 JAC54621.1 CH480817 EDW50805.1 AXCM01000793 CH902618 EDV40561.1 CH379069 EAL30969.1 CH479188 EDW40420.1 CM000363 EDX09533.1 CM002912 KMY98039.1 GFDG01000889 JAV17910.1 GANO01004079 JAB55792.1 QOIP01000006 RLU21410.1 KK107135 EZA58026.1 CM000159 EDW93724.1 GFDG01000890 JAV17909.1 GAPW01003988 JAC09610.1 KQ979568 KYN20868.1 CH478054 EAT34183.1 ADMH02000446 ETN66447.1 GGFM01004109 MBW24860.1 GGFM01003962 MBW24713.1 AJVK01014619 ATLV01019715 KE525278 KFB44566.1 CH478107 EAT33937.1 GDHF01021892 JAI30422.1 GFDF01001965 JAV12119.1 GGFJ01009084 MBW58225.1 GGFJ01009432 MBW58573.1 GGFK01010112 MBW43433.1 GEZM01034209 JAV83864.1 GL447585 EFN86349.1 CH954178 EDV50507.1 GAMD01002369 JAA99221.1 NEVH01003007 PNF40849.1 GGFK01010110 MBW43431.1 GGFK01010133 MBW43454.1 UFQS01000063 UFQT01000063 SSW98865.1 SSX19251.1 KQ977754 KYN00108.1 GGFJ01009085 MBW58226.1 GFDL01010755 JAV24290.1 GGFJ01009430 MBW58571.1 DS232001 EDS31173.1 JXJN01011623 GDHF01033240 JAI19074.1 GL435242 EFN73673.1 CH940647 EDW69637.2 CH933809 EDW18443.1 GDAI01001563 JAI16040.1 KQ976662 KYM78535.1 CH916366 EDV96524.1 PYGN01001169 PSN37013.1 GFDF01001712 JAV12372.1 GL768221 EFZ11684.1 KQ415041 KOC58909.1 GEDC01031034 JAS06264.1 KK852473 KDR23114.1 GEBQ01009050 JAT30927.1 ABLF02041084 GDKW01003170 JAI53425.1 JR041131 AEY59367.1 ACPB03005589
KPJ19106.1 KX778609 AON96594.1 ODYU01008595 SOQ52321.1 KQ458793 KPJ04985.1 AGBW02008811 OWR52505.1 JTDY01000085 KOB78972.1 KQ981864 KYN34268.1 ADTU01014817 ADTU01014818 JRES01000234 KNC33094.1 CCAG010000599 CP012525 ALC44498.1 LBMM01001065 KMQ96990.1 APCN01004335 AAAB01008960 EAA11140.2 KQ982314 KYQ57924.1 CH964095 EDW79263.1 AF199369 AE014296 AY094750 AY113387 AAF09579.1 AAF50594.1 AAM11103.1 AAM29392.1 OUUW01000002 SPP76224.1 GAKP01004331 JAC54621.1 CH480817 EDW50805.1 AXCM01000793 CH902618 EDV40561.1 CH379069 EAL30969.1 CH479188 EDW40420.1 CM000363 EDX09533.1 CM002912 KMY98039.1 GFDG01000889 JAV17910.1 GANO01004079 JAB55792.1 QOIP01000006 RLU21410.1 KK107135 EZA58026.1 CM000159 EDW93724.1 GFDG01000890 JAV17909.1 GAPW01003988 JAC09610.1 KQ979568 KYN20868.1 CH478054 EAT34183.1 ADMH02000446 ETN66447.1 GGFM01004109 MBW24860.1 GGFM01003962 MBW24713.1 AJVK01014619 ATLV01019715 KE525278 KFB44566.1 CH478107 EAT33937.1 GDHF01021892 JAI30422.1 GFDF01001965 JAV12119.1 GGFJ01009084 MBW58225.1 GGFJ01009432 MBW58573.1 GGFK01010112 MBW43433.1 GEZM01034209 JAV83864.1 GL447585 EFN86349.1 CH954178 EDV50507.1 GAMD01002369 JAA99221.1 NEVH01003007 PNF40849.1 GGFK01010110 MBW43431.1 GGFK01010133 MBW43454.1 UFQS01000063 UFQT01000063 SSW98865.1 SSX19251.1 KQ977754 KYN00108.1 GGFJ01009085 MBW58226.1 GFDL01010755 JAV24290.1 GGFJ01009430 MBW58571.1 DS232001 EDS31173.1 JXJN01011623 GDHF01033240 JAI19074.1 GL435242 EFN73673.1 CH940647 EDW69637.2 CH933809 EDW18443.1 GDAI01001563 JAI16040.1 KQ976662 KYM78535.1 CH916366 EDV96524.1 PYGN01001169 PSN37013.1 GFDF01001712 JAV12372.1 GL768221 EFZ11684.1 KQ415041 KOC58909.1 GEDC01031034 JAS06264.1 KK852473 KDR23114.1 GEBQ01009050 JAT30927.1 ABLF02041084 GDKW01003170 JAI53425.1 JR041131 AEY59367.1 ACPB03005589
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000078541 UP000005205 UP000092445 UP000037069 UP000092444 UP000092553 UP000036403 UP000092443 UP000075903 UP000075840 UP000075882 UP000078200 UP000007062 UP000075809 UP000007798 UP000075920 UP000000803 UP000268350 UP000001292 UP000075883 UP000007801 UP000001819 UP000008744 UP000000304 UP000279307 UP000075880 UP000053097 UP000002282 UP000078492 UP000008820 UP000000673 UP000092462 UP000030765 UP000076408 UP000075901 UP000008237 UP000192221 UP000008711 UP000235965 UP000078542 UP000091820 UP000002320 UP000092460 UP000000311 UP000008792 UP000009192 UP000192223 UP000095300 UP000078540 UP000001070 UP000245037 UP000075881 UP000075885 UP000095301 UP000075884 UP000053825 UP000027135 UP000007819 UP000015103
UP000078541 UP000005205 UP000092445 UP000037069 UP000092444 UP000092553 UP000036403 UP000092443 UP000075903 UP000075840 UP000075882 UP000078200 UP000007062 UP000075809 UP000007798 UP000075920 UP000000803 UP000268350 UP000001292 UP000075883 UP000007801 UP000001819 UP000008744 UP000000304 UP000279307 UP000075880 UP000053097 UP000002282 UP000078492 UP000008820 UP000000673 UP000092462 UP000030765 UP000076408 UP000075901 UP000008237 UP000192221 UP000008711 UP000235965 UP000078542 UP000091820 UP000002320 UP000092460 UP000000311 UP000008792 UP000009192 UP000192223 UP000095300 UP000078540 UP000001070 UP000245037 UP000075881 UP000075885 UP000095301 UP000075884 UP000053825 UP000027135 UP000007819 UP000015103
Pfam
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JAV8
A0A1E1WIF3
A0A2A4ITU9
A0A194RSQ6
A0A1C9EGH5
A0A2H1WGX9
+ More
A0A194QJ82 A0A212FFI3 A0A0L7LU49 A0A195F1C6 A0A158NFW3 A0A1A9ZMR6 A0A0L0CNR5 A0A1B0G6B8 A0A0M5J0W1 A0A0J7NX49 A0A1A9XTV2 A0A182VJA9 A0A182HM34 A0A182LJX1 A0A1A9UVU1 Q7Q760 A0A182V3N1 A0A151XC36 B4N3Q8 A0A182WQL9 Q9V3E3 A0A3B0J3J1 A0A034WGB2 B4HV34 A0A182MAR8 B3M4Y7 Q2M0G3 B4GRN5 B4QKB0 A0A0J9RRV2 A0A1L8EGQ1 U5ENH7 A0A3L8DLV2 A0A182IM66 A0A026WQT2 B4PK44 A0A1L8EH25 A0A023EKE2 A0A195E6R1 Q16IT0 W5JPY1 A0A2M3Z8P9 A0A2M3Z855 A0A1B0DDT5 A0A084W2X2 Q16I54 A0A0K8UUS4 A0A182Y0W2 A0A182T373 A0A1L8E0B4 A0A2M4BYQ9 A0A2M4BZV1 A0A2M4ARN5 A0A1Y1MDP8 A0A182LJL8 E2BDC1 A0A1W4UUR2 B3NFG5 T1DQN0 A0A2J7RJ36 A0A2M4ARM6 A0A2M4ARQ8 A0A336K691 A0A195CHC5 A0A2M4BYS4 A0A1A9W5R8 A0A1Q3F9P3 A0A2M4C0A4 B0WMS5 A0A1B0BBS8 A0A0K8TX66 E1ZYY3 B4LI70 B4KVR9 A0A0K8TQ72 A0A1W4W4B3 A0A1I8NXB2 A0A195B2M7 B4IY74 A0A2P8XYE1 A0A182KA93 A0A182PN78 A0A1I8N2J6 A0A1L8E1C9 E9J666 A0A182NM80 A0A0L7QJY9 A0A1B6BYY2 A0A067RTH4 A0A1B6M4T3 J9K2S1 A0A0P4VNV2 V9IFC4 T1I7I6
A0A194QJ82 A0A212FFI3 A0A0L7LU49 A0A195F1C6 A0A158NFW3 A0A1A9ZMR6 A0A0L0CNR5 A0A1B0G6B8 A0A0M5J0W1 A0A0J7NX49 A0A1A9XTV2 A0A182VJA9 A0A182HM34 A0A182LJX1 A0A1A9UVU1 Q7Q760 A0A182V3N1 A0A151XC36 B4N3Q8 A0A182WQL9 Q9V3E3 A0A3B0J3J1 A0A034WGB2 B4HV34 A0A182MAR8 B3M4Y7 Q2M0G3 B4GRN5 B4QKB0 A0A0J9RRV2 A0A1L8EGQ1 U5ENH7 A0A3L8DLV2 A0A182IM66 A0A026WQT2 B4PK44 A0A1L8EH25 A0A023EKE2 A0A195E6R1 Q16IT0 W5JPY1 A0A2M3Z8P9 A0A2M3Z855 A0A1B0DDT5 A0A084W2X2 Q16I54 A0A0K8UUS4 A0A182Y0W2 A0A182T373 A0A1L8E0B4 A0A2M4BYQ9 A0A2M4BZV1 A0A2M4ARN5 A0A1Y1MDP8 A0A182LJL8 E2BDC1 A0A1W4UUR2 B3NFG5 T1DQN0 A0A2J7RJ36 A0A2M4ARM6 A0A2M4ARQ8 A0A336K691 A0A195CHC5 A0A2M4BYS4 A0A1A9W5R8 A0A1Q3F9P3 A0A2M4C0A4 B0WMS5 A0A1B0BBS8 A0A0K8TX66 E1ZYY3 B4LI70 B4KVR9 A0A0K8TQ72 A0A1W4W4B3 A0A1I8NXB2 A0A195B2M7 B4IY74 A0A2P8XYE1 A0A182KA93 A0A182PN78 A0A1I8N2J6 A0A1L8E1C9 E9J666 A0A182NM80 A0A0L7QJY9 A0A1B6BYY2 A0A067RTH4 A0A1B6M4T3 J9K2S1 A0A0P4VNV2 V9IFC4 T1I7I6
PDB
6NU2
E-value=1.47537e-06,
Score=120
Ontologies
GO
PANTHER
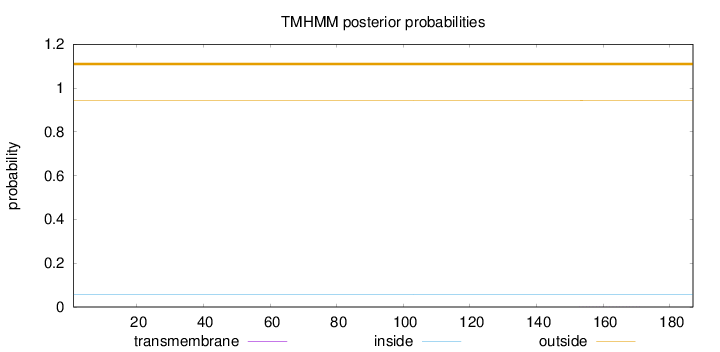
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00636
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05593
outside
1 - 187
Population Genetic Test Statistics
Pi
221.833893
Theta
189.987913
Tajima's D
0.340457
CLR
0.037125
CSRT
0.468476576171191
Interpretation
Uncertain
