Gene
KWMTBOMO05423
Pre Gene Modal
BGIBMGA006654
Annotation
PREDICTED:_protein_KIAA0100_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.465
Sequence
CDS
ATGAGCAGCCTGCGCACGTGGGGCACGCTGCTCGCCGCCGAGGAGCAGCCGCCGCCCCGGGCTGTCCGCACGGTGGTGGTGCGGCAGGGCGAGCCCTGGGGCGACGTGGAGCTGGAGCGCAGCATGATGCCGCTCAAGTGGTACTACGACCTGTGCTGCGAGATGGCGCAGTTCAGCTACGCCTTCGGGCCCTGCTGGGACCCCGTCATCGCTCACTGCAATCTCGCGTTCGACCACGTGTCCCGGCCGTCCCTGGACCCGTCCCCGCCCCTGGCCTGGTGGGACAAGGTGCGGCTGCTGATGCACGGGCGCCTCACTCTCAACTGCAGCAAGTTCACGTGCCTGCTGCACGTGTCGCTCGACCCCTACAACACCACCGAGGAGATGGAAGTCACGTGGACCGACCTCGTCTTGGACTGGACGAACGGCAAGTTGGGTTTCCTGGGCTCGCTGAGCGTGTTGGTGCGCACGGCCAGCAAGTACGACGACTGCCGCGTGCTGAGCGCCGGCCGCGTGCGCCTGGGCGTGCGCCTGGCCTGGGCGTGCGGCGCCGACCCGCGCCACCACCACGCCGTCACGCCCTGCGCGCCCACGCGCACGCCCGACTACTCCAGCAACCAGGAACACGACTCGTACCGCGCCTTCCGGTCGCAGGGCCTGGACCTGCACCTGACCCTGGAGACGAAGCCGGCCGACAGGGACGCGCCCGGACCCGTCCTGCTGCTGTACGGGAGCACGCTGCGATGGTTCGAGAGCCTGAAGCTGATCCTGTCGGGCATCACGCGGCCCACCCGCCGCGGCCGCCTGTTCAACAACGTGCGGCCCCGCAAGCCGCAGCTCTCCCGCCACTACCGGAACGTCAGCGTGTCCCTCACACTACATAATTTGCAAGTGTGCTACTGGTCGTCGTCGTCCCTGCAGCGCGGCATCGAGCTGTGCGGCGGGCGCGTGACGTGCGGCGCCAAGCACGCGCTGCGCCTGGAGCCCGCGCCCGGCCCGCTGCGCCGCCGCCCGCGCGCGCTGTGGGCCGCGCACTACCTCACCTGCGACCTCAACGACGCCGAGATCTGGCTCAAGACGCGCGCCGCCGACGACAAGATGAGTGTGGGCGGCGGCGGAGGCGACTCTCCGCAAGCGATGGAGAAGCTGTACTGTCTGAGCGTGCGGCGCGTCTCGTACACGCGCGAGGCGGGCGGGGGTGGGGGCGGGGGCGGGGGCGGGCCGGCGCACCGCGTGGCCGTGCACCACCTGCGCGGGGCCTGGACCAAGCTCAACCGGGACCTGGCCTTCGGGCTCATCGACACCTACATCAAGACGCAGCAACTGAAGAAGAATTTGTCAACAAAAGCTTTCAAGGAGGAAGAGCAGCCGGCGACCTCTCCGAAATTGCAAAGAGAAGCCGATGTCACGTCCCCGCCACCTACGACGTCACAAAGCGTGTCGAGCCAGTGCGGCGGCGGCATGCTGGCGGCGCTGGTGGCGGAGGCGGGCGGGGACGCGCCCGTCGTGTTCAGCGACGAGCTCAGCGGCGCCGACACCGCCGCGCCCGCCGCCGCCGCGCCGCACGGCCGCCTGCTGCTCGACGACGACAACTCGCACACCGCCTGCCTCGCAATTCCTCGCTGGCTCACTCTGTTCCCGTCCGCAGTGGAGCTGGTGAACTCGCAAGTGGTGCTGAAGGGGTGCGAGACGCGCGGCTACGTGATGGTGTGCGCGGCGCGGGCCGAGGTGCGGCAGCGGGTGAGCCGGCCGCCCGCGCCCGCCGCGCGCGCCTGGAGCGGCTCGCTGTCCGCCATGCAGTACTACGCCACGGTGAGCGCCGCCGACAACCACCGGCTGGACCACCACATCCAGTGGGTGGGGGTGGAGGAGATCGAGGAGCGCTGCCCGGAGATCAGCGCGCTGCCGGACGTGCCGCGGCTGGTGGGCAGCGGGCACTCTGCGGGCGGCGTCATCGGGCGCACCGTGGGGCCCAGCGCGGCCGCGGCGCAGCTGCAGCGCATCGTGTCGCGCTGCGCCTGCGAGTTCCACTACGTGTCGCTGGAGGACGCGCCGCACGCACCCGCCGCCGCGCCGCACCCCTACGACTCCTTCTCGCTCATGCACCACGACCTCGACGTCTGCACCAACTCGCTGCAGTACGCGATGCTGCTGGACGTGGTGAACAACGTGGTGCTCCACGTGGAGCCGGAGCGGCGCCGCACGCTGGAGCGCCGCGCCAGGATGCGGTTCCAGCTGCAGCTGCACCAGGACCAGGACCCGCGCCAGCAGATACACAAGCTGCAGACGCAGGTGAGAGAGTCTCTGGCCAGACTGCGTCGATTGGAGAAGGAGTACTATTTGAAGAACAGGTCCATAACGGGAAACGATCCTGTCGCTGTTGCGGAACTGAAGAGCTTGGACGATCAGGTGAACGAGTGCAAGGAGTGCGTGTGGTCGCTGGGCGAGGAGCTGGAGGCGATGGTGCGCGCGGCGCGGGAGCTGCGGCGCGAGGTGCCGGCGGGCGCGGCGCCGCACCGCTACAACGAGATCTGCTTCAAGAGCGCGCGCTGGCGGCTCACCGACGCCGACGGACAGCTGGGCATCGCCGACCTGCTGCTCACCAACTTCCTGTACACGAAGACGTCTCGTTCCGACGACTCAGTGGAGCACCAGCTGGAGGTTGGCTACGTCAGCGTCAAGAATCTGTTACCCAACGAACCCTTTCCAGAGGTGCTGGTGCCGCAGGAGCCCTCGGGCCGCGCCCCGCTGGCCCGGCGCGCCGCGCTGCGCGTGTTCTGCCGCGACCGCCCGCCCGTGGGAGGCATCGCCGTCAAGGAGCACTTCGAGGTCAACATCGTGCCCATACGGATCGGCCTTACAAAGAAGTTCTTCAATACGATGCTGAAGTTCTGTTTCCCTGAGCGGGACCCGGACTCCATAGACGAGGCGGAGGACGAGGAGGGCGCGCCCAGGCCCTCCACCGGGCTCGTCTCCACGGGCTCCAAGAAACTCAAGAAGAAGGCGAAGGACTCCAACTCCAACTTCTACGTCAAACGGGACAATAAAGATAAAGATGACGTCGAGAAGATGAAGGAGCGTGCAGAGAAGAACAAGCTGTTCATCTACATCAAGATCCCGGAGGTGCCCGTCCGCGTGTCGTACAAGGGCAGCAAGGAGAAGAACCTGGAGGACGTGCGCGACCTGCCGCTGGTGCTGCCCACGCTCGAGTACCACAACGTTACCTGGACCTGGCTCGACCTGCTGCTCGCCACCAAGAACGACACCAGGAGGGTGATCCTGTCGCAGGCCATCCGGCAGAAGCTGCAGCTGCACCGGCGCGCGGCCGCCGCGCCCGAGCCGCTGGAGGAGGACAAGGCGCGCCTGCTGCTGGGCGAGCGCACGGTCGCCGAGAACAAACCCCAGAAGAAGAGCACACCGAGCGTCTTCAAGTTTTCCAAATCGTAA
Protein
MSSLRTWGTLLAAEEQPPPRAVRTVVVRQGEPWGDVELERSMMPLKWYYDLCCEMAQFSYAFGPCWDPVIAHCNLAFDHVSRPSLDPSPPLAWWDKVRLLMHGRLTLNCSKFTCLLHVSLDPYNTTEEMEVTWTDLVLDWTNGKLGFLGSLSVLVRTASKYDDCRVLSAGRVRLGVRLAWACGADPRHHHAVTPCAPTRTPDYSSNQEHDSYRAFRSQGLDLHLTLETKPADRDAPGPVLLLYGSTLRWFESLKLILSGITRPTRRGRLFNNVRPRKPQLSRHYRNVSVSLTLHNLQVCYWSSSSLQRGIELCGGRVTCGAKHALRLEPAPGPLRRRPRALWAAHYLTCDLNDAEIWLKTRAADDKMSVGGGGGDSPQAMEKLYCLSVRRVSYTREAGGGGGGGGGGPAHRVAVHHLRGAWTKLNRDLAFGLIDTYIKTQQLKKNLSTKAFKEEEQPATSPKLQREADVTSPPPTTSQSVSSQCGGGMLAALVAEAGGDAPVVFSDELSGADTAAPAAAAPHGRLLLDDDNSHTACLAIPRWLTLFPSAVELVNSQVVLKGCETRGYVMVCAARAEVRQRVSRPPAPAARAWSGSLSAMQYYATVSAADNHRLDHHIQWVGVEEIEERCPEISALPDVPRLVGSGHSAGGVIGRTVGPSAAAAQLQRIVSRCACEFHYVSLEDAPHAPAAAPHPYDSFSLMHHDLDVCTNSLQYAMLLDVVNNVVLHVEPERRRTLERRARMRFQLQLHQDQDPRQQIHKLQTQVRESLARLRRLEKEYYLKNRSITGNDPVAVAELKSLDDQVNECKECVWSLGEELEAMVRAARELRREVPAGAAPHRYNEICFKSARWRLTDADGQLGIADLLLTNFLYTKTSRSDDSVEHQLEVGYVSVKNLLPNEPFPEVLVPQEPSGRAPLARRAALRVFCRDRPPVGGIAVKEHFEVNIVPIRIGLTKKFFNTMLKFCFPERDPDSIDEAEDEEGAPRPSTGLVSTGSKKLKKKAKDSNSNFYVKRDNKDKDDVEKMKERAEKNKLFIYIKIPEVPVRVSYKGSKEKNLEDVRDLPLVLPTLEYHNVTWTWLDLLLATKNDTRRVILSQAIRQKLQLHRRAAAAPEPLEEDKARLLLGERTVAENKPQKKSTPSVFKFSKS
Summary
Uniprot
A0A1C9EGI5
A0A194QHW3
A0A212FFJ4
A0A194RNM2
A0A088A3H9
A0A2A3E3D3
+ More
A0A0L7QS57 A0A026WXF1 A0A3L8D7G8 A0A0J7KS44 A0A195CBD4 A0A158NQG3 E2A9V2 A0A151WRW7 F4W870 E2BI45 A0A067RR67 A0A232FAS8 A0A2J7RJ51 D6WYH8 A0A023F1R5 A0A139WD06 A0A0T6AW53 A0A0V0G542 A0A224X563 A0A0M9A8A1 E0VG68 A0A151HYN7 A0A195EWK2 A0A195DUL4 A0A146MBQ1 A0A0A9XL34 A0A2S2QQA3 J9K128 A0A0C9R695 A0A1Q3FY95 A0A0K8UUE1 K7IVI0 A0A0A1WXS0 W8B5P9 A0A336L3B3 A0A336MCJ9 A0A1A9XV08 A0A1A9V913 B4KXH9 A0A1B0BI36 W4VRT9 A0A1A9X0V8 A0A0L0CL85 A0A1B0ET94 A0A182H5I7 A0A1B6LCV2 A0A1W4W147 A0A1I8M9M2 A0A1I8QBK5 A0A182V0U4 B4LFV3 T1PAZ2 A0A182HM39 B3NBT0 Q9VZS7 A0A0J9RN32 B3M5U2 A0A084W2W2 B4N3Q3 A0A226ET55 A0A3B0JRA2 A0A0M4EJ32 A0A182LJL7 Q7PNQ7 A0A182TGF5 A0A182U3U6 A0A182XIK4 A0A034VXT7 A0A1D2NFL7 E9FY09 A0A164TZK4 A0A0N8D3P6 A0A0P6B0W9 A0A0P5YXG6 A0A0P6H2D3 A0A0P5C930 A0A0P5PDV3 A0A1B6F5J7 A0A3R7P532 A0A0P6DY67 A0A1B0AES6 A0A087T0W7 Q16IU0 A0A2J7RJ58 B4HTN4 B4QP53 B4IX55 A0A1J1I3B7 A0A2H8TN67 A0A2R5LDS7 L7MLL4
A0A0L7QS57 A0A026WXF1 A0A3L8D7G8 A0A0J7KS44 A0A195CBD4 A0A158NQG3 E2A9V2 A0A151WRW7 F4W870 E2BI45 A0A067RR67 A0A232FAS8 A0A2J7RJ51 D6WYH8 A0A023F1R5 A0A139WD06 A0A0T6AW53 A0A0V0G542 A0A224X563 A0A0M9A8A1 E0VG68 A0A151HYN7 A0A195EWK2 A0A195DUL4 A0A146MBQ1 A0A0A9XL34 A0A2S2QQA3 J9K128 A0A0C9R695 A0A1Q3FY95 A0A0K8UUE1 K7IVI0 A0A0A1WXS0 W8B5P9 A0A336L3B3 A0A336MCJ9 A0A1A9XV08 A0A1A9V913 B4KXH9 A0A1B0BI36 W4VRT9 A0A1A9X0V8 A0A0L0CL85 A0A1B0ET94 A0A182H5I7 A0A1B6LCV2 A0A1W4W147 A0A1I8M9M2 A0A1I8QBK5 A0A182V0U4 B4LFV3 T1PAZ2 A0A182HM39 B3NBT0 Q9VZS7 A0A0J9RN32 B3M5U2 A0A084W2W2 B4N3Q3 A0A226ET55 A0A3B0JRA2 A0A0M4EJ32 A0A182LJL7 Q7PNQ7 A0A182TGF5 A0A182U3U6 A0A182XIK4 A0A034VXT7 A0A1D2NFL7 E9FY09 A0A164TZK4 A0A0N8D3P6 A0A0P6B0W9 A0A0P5YXG6 A0A0P6H2D3 A0A0P5C930 A0A0P5PDV3 A0A1B6F5J7 A0A3R7P532 A0A0P6DY67 A0A1B0AES6 A0A087T0W7 Q16IU0 A0A2J7RJ58 B4HTN4 B4QP53 B4IX55 A0A1J1I3B7 A0A2H8TN67 A0A2R5LDS7 L7MLL4
Pubmed
26354079
22118469
24508170
30249741
21347285
20798317
+ More
21719571 24845553 28648823 18362917 19820115 25474469 20566863 26823975 25401762 20075255 25830018 24495485 17994087 26108605 26483478 25315136 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 20966253 12364791 25348373 27289101 21292972 17510324 25576852
21719571 24845553 28648823 18362917 19820115 25474469 20566863 26823975 25401762 20075255 25830018 24495485 17994087 26108605 26483478 25315136 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 24438588 20966253 12364791 25348373 27289101 21292972 17510324 25576852
EMBL
KX778609
AON96595.1
KQ458793
KPJ04984.1
AGBW02008811
OWR52504.1
+ More
KQ459984 KPJ19107.1 KZ288403 PBC26208.1 KQ414758 KOC61463.1 KK107078 EZA60508.1 QOIP01000011 RLU16427.1 LBMM01003812 KMQ93106.1 KQ978023 KYM98010.1 ADTU01023310 ADTU01023311 GL437958 EFN69785.1 KQ982803 KYQ50553.1 GL887898 EGI69565.1 GL448450 EFN84616.1 KK852473 KDR23115.1 NNAY01000576 OXU27590.1 NEVH01003007 PNF40865.1 KQ971362 EFA07873.1 GBBI01003265 JAC15447.1 KYB25782.1 LJIG01022721 KRT79061.1 GECL01002938 JAP03186.1 GFTR01008789 JAW07637.1 KQ435724 KOX78506.1 DS235132 EEB12374.1 KQ976723 KYM76690.1 KQ981948 KYN32655.1 KQ980322 KYN16568.1 GDHC01002453 JAQ16176.1 GBHO01025769 JAG17835.1 GGMS01010507 MBY79710.1 ABLF02041084 GBYB01011834 JAG81601.1 GFDL01002490 JAV32555.1 GDHF01022171 JAI30143.1 GBXI01011079 JAD03213.1 GAMC01017941 GAMC01017939 GAMC01017937 JAB88616.1 UFQS01000930 UFQT01000930 SSX07814.1 SSX28048.1 UFQS01000941 UFQT01000941 SSX07888.1 SSX28122.1 CH933809 EDW18665.2 JXJN01014762 GANO01000139 JAB59732.1 JRES01000234 KNC33123.1 CCAG010004390 JXUM01026475 JXUM01026476 KQ560757 KXJ81002.1 GEBQ01018456 JAT21521.1 CH940647 EDW69330.2 KA645320 AFP59949.1 APCN01004337 CH954178 EDV50747.2 KQS43483.1 AE014296 AAF47740.2 ALI30451.1 CM002912 KMY97303.1 CH902618 EDV39632.2 ATLV01019709 ATLV01019710 KE525278 KFB44556.1 CH964095 EDW79258.1 LNIX01000002 OXA60410.1 OUUW01000002 SPP76219.1 CP012525 ALC42922.1 AAAB01008960 EAA11765.3 GAKP01012594 JAC46358.1 LJIJ01000056 ODN04060.1 GL732526 EFX88195.1 LRGB01001663 KZS10916.1 GDIP01071879 JAM31836.1 GDIP01021606 JAM82109.1 GDIP01053230 JAM50485.1 GDIQ01025501 JAN69236.1 GDIP01177915 JAJ45487.1 GDIQ01132212 JAL19514.1 GECZ01024233 JAS45536.1 QCYY01001749 ROT75648.1 GDIQ01085681 JAN09056.1 KK112883 KFM58756.1 CH478054 EAT34174.1 PNF40866.1 CH480817 EDW50305.1 CM000363 EDX09048.1 CH916366 EDV96361.1 CVRI01000040 CRK94800.1 GFXV01003808 MBW15613.1 GGLE01003503 MBY07629.1 GACK01000980 JAA64054.1
KQ459984 KPJ19107.1 KZ288403 PBC26208.1 KQ414758 KOC61463.1 KK107078 EZA60508.1 QOIP01000011 RLU16427.1 LBMM01003812 KMQ93106.1 KQ978023 KYM98010.1 ADTU01023310 ADTU01023311 GL437958 EFN69785.1 KQ982803 KYQ50553.1 GL887898 EGI69565.1 GL448450 EFN84616.1 KK852473 KDR23115.1 NNAY01000576 OXU27590.1 NEVH01003007 PNF40865.1 KQ971362 EFA07873.1 GBBI01003265 JAC15447.1 KYB25782.1 LJIG01022721 KRT79061.1 GECL01002938 JAP03186.1 GFTR01008789 JAW07637.1 KQ435724 KOX78506.1 DS235132 EEB12374.1 KQ976723 KYM76690.1 KQ981948 KYN32655.1 KQ980322 KYN16568.1 GDHC01002453 JAQ16176.1 GBHO01025769 JAG17835.1 GGMS01010507 MBY79710.1 ABLF02041084 GBYB01011834 JAG81601.1 GFDL01002490 JAV32555.1 GDHF01022171 JAI30143.1 GBXI01011079 JAD03213.1 GAMC01017941 GAMC01017939 GAMC01017937 JAB88616.1 UFQS01000930 UFQT01000930 SSX07814.1 SSX28048.1 UFQS01000941 UFQT01000941 SSX07888.1 SSX28122.1 CH933809 EDW18665.2 JXJN01014762 GANO01000139 JAB59732.1 JRES01000234 KNC33123.1 CCAG010004390 JXUM01026475 JXUM01026476 KQ560757 KXJ81002.1 GEBQ01018456 JAT21521.1 CH940647 EDW69330.2 KA645320 AFP59949.1 APCN01004337 CH954178 EDV50747.2 KQS43483.1 AE014296 AAF47740.2 ALI30451.1 CM002912 KMY97303.1 CH902618 EDV39632.2 ATLV01019709 ATLV01019710 KE525278 KFB44556.1 CH964095 EDW79258.1 LNIX01000002 OXA60410.1 OUUW01000002 SPP76219.1 CP012525 ALC42922.1 AAAB01008960 EAA11765.3 GAKP01012594 JAC46358.1 LJIJ01000056 ODN04060.1 GL732526 EFX88195.1 LRGB01001663 KZS10916.1 GDIP01071879 JAM31836.1 GDIP01021606 JAM82109.1 GDIP01053230 JAM50485.1 GDIQ01025501 JAN69236.1 GDIP01177915 JAJ45487.1 GDIQ01132212 JAL19514.1 GECZ01024233 JAS45536.1 QCYY01001749 ROT75648.1 GDIQ01085681 JAN09056.1 KK112883 KFM58756.1 CH478054 EAT34174.1 PNF40866.1 CH480817 EDW50305.1 CM000363 EDX09048.1 CH916366 EDV96361.1 CVRI01000040 CRK94800.1 GFXV01003808 MBW15613.1 GGLE01003503 MBY07629.1 GACK01000980 JAA64054.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000005203
UP000242457
UP000053825
+ More
UP000053097 UP000279307 UP000036403 UP000078542 UP000005205 UP000000311 UP000075809 UP000007755 UP000008237 UP000027135 UP000215335 UP000235965 UP000007266 UP000053105 UP000009046 UP000078540 UP000078541 UP000078492 UP000007819 UP000002358 UP000092443 UP000078200 UP000009192 UP000092460 UP000091820 UP000037069 UP000092444 UP000069940 UP000249989 UP000192221 UP000095301 UP000095300 UP000075903 UP000008792 UP000075840 UP000008711 UP000000803 UP000007801 UP000030765 UP000007798 UP000198287 UP000268350 UP000092553 UP000075882 UP000007062 UP000075902 UP000076407 UP000094527 UP000000305 UP000076858 UP000283509 UP000092445 UP000054359 UP000008820 UP000001292 UP000000304 UP000001070 UP000183832
UP000053097 UP000279307 UP000036403 UP000078542 UP000005205 UP000000311 UP000075809 UP000007755 UP000008237 UP000027135 UP000215335 UP000235965 UP000007266 UP000053105 UP000009046 UP000078540 UP000078541 UP000078492 UP000007819 UP000002358 UP000092443 UP000078200 UP000009192 UP000092460 UP000091820 UP000037069 UP000092444 UP000069940 UP000249989 UP000192221 UP000095301 UP000095300 UP000075903 UP000008792 UP000075840 UP000008711 UP000000803 UP000007801 UP000030765 UP000007798 UP000198287 UP000268350 UP000092553 UP000075882 UP000007062 UP000075902 UP000076407 UP000094527 UP000000305 UP000076858 UP000283509 UP000092445 UP000054359 UP000008820 UP000001292 UP000000304 UP000001070 UP000183832
Interpro
ProteinModelPortal
A0A1C9EGI5
A0A194QHW3
A0A212FFJ4
A0A194RNM2
A0A088A3H9
A0A2A3E3D3
+ More
A0A0L7QS57 A0A026WXF1 A0A3L8D7G8 A0A0J7KS44 A0A195CBD4 A0A158NQG3 E2A9V2 A0A151WRW7 F4W870 E2BI45 A0A067RR67 A0A232FAS8 A0A2J7RJ51 D6WYH8 A0A023F1R5 A0A139WD06 A0A0T6AW53 A0A0V0G542 A0A224X563 A0A0M9A8A1 E0VG68 A0A151HYN7 A0A195EWK2 A0A195DUL4 A0A146MBQ1 A0A0A9XL34 A0A2S2QQA3 J9K128 A0A0C9R695 A0A1Q3FY95 A0A0K8UUE1 K7IVI0 A0A0A1WXS0 W8B5P9 A0A336L3B3 A0A336MCJ9 A0A1A9XV08 A0A1A9V913 B4KXH9 A0A1B0BI36 W4VRT9 A0A1A9X0V8 A0A0L0CL85 A0A1B0ET94 A0A182H5I7 A0A1B6LCV2 A0A1W4W147 A0A1I8M9M2 A0A1I8QBK5 A0A182V0U4 B4LFV3 T1PAZ2 A0A182HM39 B3NBT0 Q9VZS7 A0A0J9RN32 B3M5U2 A0A084W2W2 B4N3Q3 A0A226ET55 A0A3B0JRA2 A0A0M4EJ32 A0A182LJL7 Q7PNQ7 A0A182TGF5 A0A182U3U6 A0A182XIK4 A0A034VXT7 A0A1D2NFL7 E9FY09 A0A164TZK4 A0A0N8D3P6 A0A0P6B0W9 A0A0P5YXG6 A0A0P6H2D3 A0A0P5C930 A0A0P5PDV3 A0A1B6F5J7 A0A3R7P532 A0A0P6DY67 A0A1B0AES6 A0A087T0W7 Q16IU0 A0A2J7RJ58 B4HTN4 B4QP53 B4IX55 A0A1J1I3B7 A0A2H8TN67 A0A2R5LDS7 L7MLL4
A0A0L7QS57 A0A026WXF1 A0A3L8D7G8 A0A0J7KS44 A0A195CBD4 A0A158NQG3 E2A9V2 A0A151WRW7 F4W870 E2BI45 A0A067RR67 A0A232FAS8 A0A2J7RJ51 D6WYH8 A0A023F1R5 A0A139WD06 A0A0T6AW53 A0A0V0G542 A0A224X563 A0A0M9A8A1 E0VG68 A0A151HYN7 A0A195EWK2 A0A195DUL4 A0A146MBQ1 A0A0A9XL34 A0A2S2QQA3 J9K128 A0A0C9R695 A0A1Q3FY95 A0A0K8UUE1 K7IVI0 A0A0A1WXS0 W8B5P9 A0A336L3B3 A0A336MCJ9 A0A1A9XV08 A0A1A9V913 B4KXH9 A0A1B0BI36 W4VRT9 A0A1A9X0V8 A0A0L0CL85 A0A1B0ET94 A0A182H5I7 A0A1B6LCV2 A0A1W4W147 A0A1I8M9M2 A0A1I8QBK5 A0A182V0U4 B4LFV3 T1PAZ2 A0A182HM39 B3NBT0 Q9VZS7 A0A0J9RN32 B3M5U2 A0A084W2W2 B4N3Q3 A0A226ET55 A0A3B0JRA2 A0A0M4EJ32 A0A182LJL7 Q7PNQ7 A0A182TGF5 A0A182U3U6 A0A182XIK4 A0A034VXT7 A0A1D2NFL7 E9FY09 A0A164TZK4 A0A0N8D3P6 A0A0P6B0W9 A0A0P5YXG6 A0A0P6H2D3 A0A0P5C930 A0A0P5PDV3 A0A1B6F5J7 A0A3R7P532 A0A0P6DY67 A0A1B0AES6 A0A087T0W7 Q16IU0 A0A2J7RJ58 B4HTN4 B4QP53 B4IX55 A0A1J1I3B7 A0A2H8TN67 A0A2R5LDS7 L7MLL4
Ontologies
GO
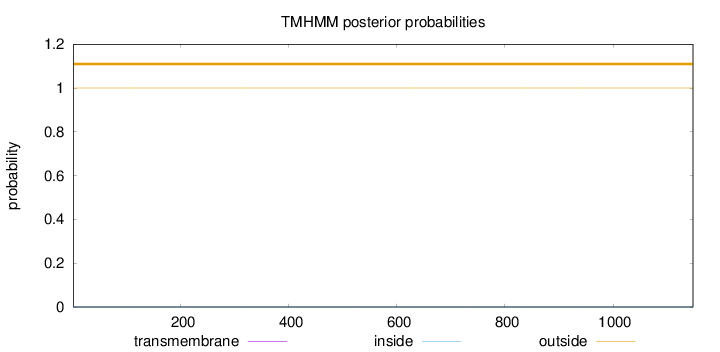
Topology
Length:
1148
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0048
Exp number, first 60 AAs:
0.00049
Total prob of N-in:
0.00015
outside
1 - 1148
Population Genetic Test Statistics
Pi
177.423528
Theta
154.30673
Tajima's D
0.409162
CLR
1.016264
CSRT
0.485325733713314
Interpretation
Uncertain
