Gene
KWMTBOMO05421 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006655
Annotation
PREDICTED:_integrin_alpha-PS2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.523 Nuclear Reliability : 1.772
Sequence
CDS
ATGCCGAAAATACCGGACGAAGGTCAGCTGATGTCACAGACGCTATACGCCCGGCCGGCCCTTATAGCTACGCTAGAGTCAAGTCCTAGGTACGACGACTCTTACATGGGATACTCTATAACCGTTGGTGATTTTGCTGGACAAGGCATTCAGAGTGTTGCTGTGGGTATGCCTAGAGGTGCTGACTTGAGGGGTTTGGTGGTACTCTACACTTGGGAGCTCCAGAACATCAAGAATATATCCGGCTCTCAGATCGGAGCATACTTCGGTTACAGTCTCGCTTCTGGTGACATTGACGGTGATGGAACCGATGACGTCATAGTAGGAGCTCCAATGTTCACCAAGCCGAGAAGCGACGGATACGAACACGGAAGAATCTATGTTATTTACCAGGGAACTGACAGGTCGTTCTCAAGAAGTCACGCAAGGACCGGCGAGGTCTCCCAGGGCAGGTTCGGGTTGGCGGTAACGTCACTCGGTGACATCAACTATGACGGATTCGGCGATATCGCGGTGGGAGCTCCATACGGCGGAGAGAAAGGCCGGGGCGTGGTCTACATCTACCACGGAAGCGAAGTCGGCATATTGGAGAAGTACTCGCAGGCCATCACCGCGGAGGAGATCTCGCCGACCTTGAGCACGTTCGGCTTCTCGCTGTCCGGCGGAGTGGACCTCGACAACAACAACTACACCGATTTGGCAGTCGGAGCCTATAAGTCCAACAGTGTTGTGTTTTTAAAGTCTCGTCCCGTCGTCAAGGTGAGTGCCGAAGTGAAGTTCATGGGCGAGAGCAAGCTCATCTCGCTCACGGACAAGAAGTGCCATCTCAGTAACGGCACGGAGGTCGCGTGCGCCCAGGTCATGTTCTGTCTCACCTACACCGGGGTCAACGTGGACCAGCAGATAAATTTCGAGGTGACTTTAGACCTGGATTCGCGTCAGACGACCAGCAAGCGACTGTTCTTAGCTGAAACCAGAGAGACGACATACAAGACCCACATCATGCTGACGCAGGGACTGCAGGAGTGCAAAGATGTCGTTGTCTACTTAGATGAGGAGATCCGCGACAAGCTGACGCCCATAGAGGTGAAGCTGTCGTACGAGCTGGTGCCGCAGCCGTCGGGCGCCGCCGTCCCGCCCGTGCTGGACCGCACGCGCGCCGCCGCGCACTCCGACGCGCTCCACATACAGAAGAACTGCGGACCCGACAACATCTGCATCCCCGACCTCAGGATGTCCGCCGCCACCCCGATCGTGAACTACGTTCTCGGCTCCGGAGAAAACTTGAACATCGATGTGAAAGTGGAGAACTCCGGCGAGGACGCCTTCGAAGCTGCCTACTACCTGCTGATTCCTGCCGGAGTTACATACGCGAAGACCGAACGTTTAGACAAGGACAATATAGTGACTCCTATATACTGTTCTATAGAGAAGAGGGAATCCGACGGGAACTCTACTTTGAAGTGTGATATAGGCAACCCCATGGCCAGTGGACAGAAGGTGAACTTCCGCGTGATCCTCGAAGTGGACGCGCTCGTGACGTCACTGAACTTCTACATGGAAGCGAACTCCACCAACCCCGAGCAAGGAACCGATTACGACAACGTCATGAACATGAACATCGGCGTCATCATCAGAGCACAGCTCTCTGTCATCGGGACCTCAGACCCGCCCGAGCTCCATTACAACGCGTCGGTATACGAAGCGACCGACCTGAAGGACGACACCAAGCTGGGCCCCCCCGTGATCCACAAGTACAGCATCAAGAACGAGGGGCCCTTCACCGTAGACGAAGCCGAGATGTACTTCATGTGGCCCTATCAGACTCTTGAAGGCGAAAACCTGATGTACATGCTGGTGCAGCCGCAGTGGCTCGGGAACATCAAGTGCAACGTGGCGCGACACATCAACACAGAGAACCTCTTCGTCCAGAATCCCTACATGCCTCTGCTCGAAAAGGAAAGAGAAGCAATGGCGAGCACCAACATGTACACTGAAGCACAGCTCGTCGGTGGCTTCGGATATTACGGGCAGACGCAATGGGAGCAGCATTCCACCAGCGGCGGTACAGTGACGTCACAATCTGGAGGTTTCGTGTCAGGTGGACAACAGGGATCGTCCACCCAAACTGGCCCCGCCAATTCCGGTGGTCAGCTTAGTAGCGGTCAAACTGCTGGTGGTCAGTTCACTCACACTTCTGGAGGTCAATACAGCGGAGGTCAAACTGCTGGTGGGCAGTTTAGTCAATCTTCCGGAGGTCCATACGGTGGAGATCACAGTGCTGGTGGGCAGTTTAGTCAAACTTCCGGAGGTCAGTATAGTGGAGGTCAAGAACAGCAATCTGGACAACATCAGTTCAGTCAGGGAGGACAGAACGGCGGAACTACTTATACATACAAGTGGACGAATGGACAAGGTCTGACTGTTGAAGAACAGGAGAAAATCAAGAAAAGTCTTGAAGCCTTCGCACAAGGAAACGGGCAGGGCGGTTTCACCCAGGGTGGCTTCACTCAAGGAGCCGACGGACAGATCTATGTCGATGGAGGAAGAGTCGTTCGCATTAGGAATATAACGACAGTTTACGACGCAAATAACAACACCATATTCCAGACTGAGACTAGCACGGAATACGGAAAACTTGGGCATGAAGGTGAAGCTGGAAGTTCATTTAATACATACGTTAATAACCAAGGAGGACAGCAACACAGCGGCTCGTTCGCTCACGGCTCTGGTGGTTACGTCCAAGGTTCGGGCCAAACTCAGACCCAAGGTGGACAAAATTGGCAAGGCTCTGTTCAAGGAAATAGTCAGTTCTCTCAAGGCTCAGGATCTGGACAAGGCTTTGTCCATGGATCCTGGGGAACCACAGAGACAGTTAACCCTGACCTTGTGAAAGCCGGGTCGTCGACTTTCAGAGGAGCCTCAACAGTCACTGTAGTGGGTGACGAAGAAGACAACGAAGGACTTGGTTTTGGAGCTTTCGCTGCTTCTAACCCTAACAATGAGTTCAAATACGCGATCGGTGACGTAACCAACGCCGGCAGCGGACAAGCTGGAAGTCGCACCGGCACTGCTGGTGGTAGTTACCAATCTGGCGGGGCTTATCAAGCTGGAGGAGCCGGGCATCAAGCTGGAGGAGGTGCTTATCTGTCTGGTGGAGGCAGTTACCGAACTGGAGGTTCTGCTTATCAGTCTGGGGGGAGCGCACAGGATGGAAGCCGATCGTGGAGCAGTGGATATTCTTATTCTTCTCAATCAGGAGGTGGCTCTTCATACAACAGCCATTCTGGAAATTATGGCTCCACTAGTAGCCGAGAAAACACCAGGAGAAGGAGACAGGTCGATCAGTCACAAGTTGACAAGCAACTCAAGGACATCTTAACCAACTGTCAGGAGAAATACAAATGTGAAGTGATGAGGTGCACCACGGGCAGGCTGCTGAAGGGCCAGGAGGTGTGGGTGGCGCTGCGGTCCAGGATCAACGCAACTGTACTTAACGAGATCTCGAAGGAGCGGCCGATAGTGCTGTCGTCGCTGTGCGCGGCGCGCGTGTCGCGGCTGGGCGGCGGCGCGGCGGGCGGCGCGGGGGCGGGCTGGGCGCGGGGGGAGGCGCGCACGTCGCTGAGCCCGCAGCTGGGCGCGCGCGGGGGCGGCGTGCCGCTGTGGGTCATCATCCTCGCCGCCATCATCGGGGCGCTGCTGCTGCTCCTGCTCATCTTCGCGCTCTACAAATGCGGATTCTTCAAGCGCAACCGTCCCTCCGACCACACCGAGCGCCAGCCGCTGAACGGACGTGACGAACACCTTTGA
Protein
MPKIPDEGQLMSQTLYARPALIATLESSPRYDDSYMGYSITVGDFAGQGIQSVAVGMPRGADLRGLVVLYTWELQNIKNISGSQIGAYFGYSLASGDIDGDGTDDVIVGAPMFTKPRSDGYEHGRIYVIYQGTDRSFSRSHARTGEVSQGRFGLAVTSLGDINYDGFGDIAVGAPYGGEKGRGVVYIYHGSEVGILEKYSQAITAEEISPTLSTFGFSLSGGVDLDNNNYTDLAVGAYKSNSVVFLKSRPVVKVSAEVKFMGESKLISLTDKKCHLSNGTEVACAQVMFCLTYTGVNVDQQINFEVTLDLDSRQTTSKRLFLAETRETTYKTHIMLTQGLQECKDVVVYLDEEIRDKLTPIEVKLSYELVPQPSGAAVPPVLDRTRAAAHSDALHIQKNCGPDNICIPDLRMSAATPIVNYVLGSGENLNIDVKVENSGEDAFEAAYYLLIPAGVTYAKTERLDKDNIVTPIYCSIEKRESDGNSTLKCDIGNPMASGQKVNFRVILEVDALVTSLNFYMEANSTNPEQGTDYDNVMNMNIGVIIRAQLSVIGTSDPPELHYNASVYEATDLKDDTKLGPPVIHKYSIKNEGPFTVDEAEMYFMWPYQTLEGENLMYMLVQPQWLGNIKCNVARHINTENLFVQNPYMPLLEKEREAMASTNMYTEAQLVGGFGYYGQTQWEQHSTSGGTVTSQSGGFVSGGQQGSSTQTGPANSGGQLSSGQTAGGQFTHTSGGQYSGGQTAGGQFSQSSGGPYGGDHSAGGQFSQTSGGQYSGGQEQQSGQHQFSQGGQNGGTTYTYKWTNGQGLTVEEQEKIKKSLEAFAQGNGQGGFTQGGFTQGADGQIYVDGGRVVRIRNITTVYDANNNTIFQTETSTEYGKLGHEGEAGSSFNTYVNNQGGQQHSGSFAHGSGGYVQGSGQTQTQGGQNWQGSVQGNSQFSQGSGSGQGFVHGSWGTTETVNPDLVKAGSSTFRGASTVTVVGDEEDNEGLGFGAFAASNPNNEFKYAIGDVTNAGSGQAGSRTGTAGGSYQSGGAYQAGGAGHQAGGGAYLSGGGSYRTGGSAYQSGGSAQDGSRSWSSGYSYSSQSGGGSSYNSHSGNYGSTSSRENTRRRRQVDQSQVDKQLKDILTNCQEKYKCEVMRCTTGRLLKGQEVWVALRSRINATVLNEISKERPIVLSSLCAARVSRLGGGAAGGAGAGWARGEARTSLSPQLGARGGGVPLWVIILAAIIGALLLLLLIFALYKCGFFKRNRPSDHTERQPLNGRDEHL
Summary
Subunit
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the integrin alpha chain family.
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF69179
SSF69179
Gene 3D
ProteinModelPortal
PDB
6AVU
E-value=4.02268e-77,
Score=738
Ontologies
GO
Topology
Subcellular location
Membrane
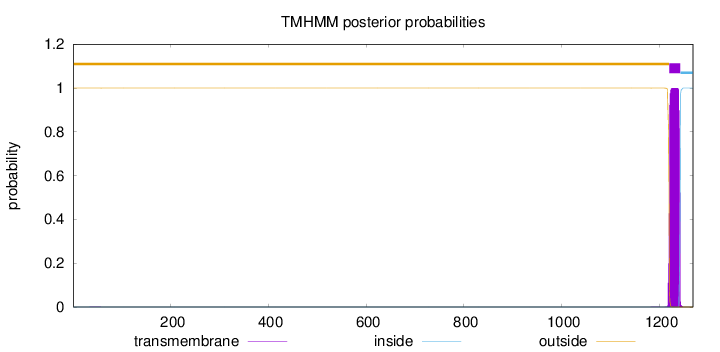
Length:
1269
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.91827
Exp number, first 60 AAs:
0.00706
Total prob of N-in:
0.00036
outside
1 - 1220
TMhelix
1221 - 1243
inside
1244 - 1269
Population Genetic Test Statistics
Pi
227.465163
Theta
180.993677
Tajima's D
0.700632
CLR
0.535834
CSRT
0.575471226438678
Interpretation
Uncertain
