Gene
KWMTBOMO05417
Pre Gene Modal
BGIBMGA006657
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_glutamate_receptor_ionotropic?_delta-2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.095
Sequence
CDS
ATGTTGCTCGTCGTGTCATTACCGAAACTGCCAACAGGCTGGTCTATTCGATTCCTGACTGGCTGGTATTGGTTGTACTGTATATTACTGGTGGTTTCGTACCGAGCAAGCATGACCGCTATCCTGGCTAACCCAGCGCCAAGAGTGACGATCGACACGTTAGTCGAATTGGCCGCCAGTAAACTTACTTGTGGAGGTTGGGGAATCGAAACTAAGAATTTTTTTCAAGATTCCTTGGATGAAATTGGTCAAAAAATTTCTGACAGATTCGAAATTAGTAACGACCCTAATATTGCGGCTGATAAAGTCGCTCAGGGTACATTTGCGTATTATGACAATAAAAACTTCCTAAAATACATCACCGTTAGAAGACAAAATGGTTTTATTATGGAAACTATTGATAACACAACTAATTTTACCAGCATTAGCACTAAAAGCAACAATGAGAGAAATCTCCATATCATGTCTGATTGCGTTGTGAACATACCTATCTCAATTGGATTTCACAAGAATTCTCCACTTAAGCCCTTAACTGATATATACATCACGAGAATAGTTGAGGTTGGTTTGGTTGAAAAGTGGCTGAATGATGCCATGTACACAATCAAAACTTTAGAAACAAACGAGGAGGAGATCAAAGCTTTAATGAACTTGAAGAAGCTTTATGGAGCCTTCATCGCATTAGCCATCGGCTACTTCTTGAGCGTGATGTGCCTAATTGGAGAGCTGGCCCACTGGAATTGTGTTGTGAAGAAAGACCCAAACTATGACAAATATGCTTTACACAAATATTATGAGAAAATAAACAAAAAGCAGTAG
Protein
MLLVVSLPKLPTGWSIRFLTGWYWLYCILLVVSYRASMTAILANPAPRVTIDTLVELAASKLTCGGWGIETKNFFQDSLDEIGQKISDRFEISNDPNIAADKVAQGTFAYYDNKNFLKYITVRRQNGFIMETIDNTTNFTSISTKSNNERNLHIMSDCVVNIPISIGFHKNSPLKPLTDIYITRIVEVGLVEKWLNDAMYTIKTLETNEEEIKALMNLKKLYGAFIALAIGYFLSVMCLIGELAHWNCVVKKDPNYDKYALHKYYEKINKKQ
Summary
Uniprot
H9JAW2
A0A075TD94
A0A0K8TVJ6
A0A2A4J5W4
A0A386H8Z8
A0A0L7L303
+ More
A0A345BF47 A0A1B3B704 A0A0F7QK03 A0A1B3P5I4 A0A2H1VUJ7 E5FIA7 A0A2K8GKZ5 A0A2K8GL85 A0A1Y9TJT5 A0A194QNB4 A0A194RMR1 A0A140G9I2 A0A3S2LVR8 A0A1C9EGI0 A0A212EVL8 A0A2P8XM32 A0A0U5ARX4 A0A067RDK9 D6X0V7 J9KAI3 A0A1B6IQG3 A0A310SCX2 A0A1W4XI02 T1H8I6 E0VMW0 A0A2R7VZ94 A0A026X2P0 A0A3L8DNP4 A0A195F448 A0A0A9WWV2 A0A0M8ZUV9 A0A310S941 F4W4Y1 A0A151XEJ6 A0A151JLH6 A0A2A3EKI7 A0A087ZVT3 A0A195CF85 A0A195BNX6 A0A0L7QJU4 A0A154PSX2 Q17BA7 A0A336MB17 E2A0W6 A0A3L9LSY2 A0A1S4F9D5 A0A1W6L170 A0A182H107 B0W521 A0A182W7U8 A0A182MDX7 A0A232FKC0 A0A182Q3D6 A0A1J1ITH8 A0A182YGS4 K7IQ73 A0A182RVK5 A0A0M4EII4 A0A1A9VXX5 A0A0L0C9S2 A0A1S4GWS3 Q7Q3M6 A0A1W4W5W6 A0A1I8NEI8 A0A0J9RTM5 B4MKM1 A0A1A9XRY2 A0A1B0BS36 B3M3Y7 A0A1B0FPF1 B4L1C0 A0A0J9RTA8 A0A3Q0IMF8 A0A1I8PRW8 B3NGT8 A0A1A9ZKB3 B4PF26 Q9VTH3 B4J2L4 A0A3B0JWG1 X2JCB2 B4QPS2 Q2LZ47 B4GZM0 A0A1A9WUV8 A0A2K6VB67 A0A182F4J2 B4HEC2 A0A084VNZ9 A0A1B0EYB4 B4LCM8 A0A0J7KKB3
A0A345BF47 A0A1B3B704 A0A0F7QK03 A0A1B3P5I4 A0A2H1VUJ7 E5FIA7 A0A2K8GKZ5 A0A2K8GL85 A0A1Y9TJT5 A0A194QNB4 A0A194RMR1 A0A140G9I2 A0A3S2LVR8 A0A1C9EGI0 A0A212EVL8 A0A2P8XM32 A0A0U5ARX4 A0A067RDK9 D6X0V7 J9KAI3 A0A1B6IQG3 A0A310SCX2 A0A1W4XI02 T1H8I6 E0VMW0 A0A2R7VZ94 A0A026X2P0 A0A3L8DNP4 A0A195F448 A0A0A9WWV2 A0A0M8ZUV9 A0A310S941 F4W4Y1 A0A151XEJ6 A0A151JLH6 A0A2A3EKI7 A0A087ZVT3 A0A195CF85 A0A195BNX6 A0A0L7QJU4 A0A154PSX2 Q17BA7 A0A336MB17 E2A0W6 A0A3L9LSY2 A0A1S4F9D5 A0A1W6L170 A0A182H107 B0W521 A0A182W7U8 A0A182MDX7 A0A232FKC0 A0A182Q3D6 A0A1J1ITH8 A0A182YGS4 K7IQ73 A0A182RVK5 A0A0M4EII4 A0A1A9VXX5 A0A0L0C9S2 A0A1S4GWS3 Q7Q3M6 A0A1W4W5W6 A0A1I8NEI8 A0A0J9RTM5 B4MKM1 A0A1A9XRY2 A0A1B0BS36 B3M3Y7 A0A1B0FPF1 B4L1C0 A0A0J9RTA8 A0A3Q0IMF8 A0A1I8PRW8 B3NGT8 A0A1A9ZKB3 B4PF26 Q9VTH3 B4J2L4 A0A3B0JWG1 X2JCB2 B4QPS2 Q2LZ47 B4GZM0 A0A1A9WUV8 A0A2K6VB67 A0A182F4J2 B4HEC2 A0A084VNZ9 A0A1B0EYB4 B4LCM8 A0A0J7KKB3
Pubmed
19121390
25027790
26017144
26227816
29727827
25803580
+ More
27538507 21091811 28736530 29375398 26354079 27004525 22118469 29403074 26760975 24845553 18362917 19820115 20566863 24508170 30249741 25401762 21719571 17510324 20798317 26483478 28648823 25244985 20075255 26108605 12364791 25315136 22936249 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 20920257 24438588
27538507 21091811 28736530 29375398 26354079 27004525 22118469 29403074 26760975 24845553 18362917 19820115 20566863 24508170 30249741 25401762 21719571 17510324 20798317 26483478 28648823 25244985 20075255 26108605 12364791 25315136 22936249 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 20920257 24438588
EMBL
BABH01008325
BABH01008326
BABH01008327
KF768727
AIG51921.1
GCVX01000135
+ More
JAI18095.1 NWSH01003171 PCG66830.1 MG546654 AYD42277.1 JTDY01003244 KOB69888.1 MG820700 AXF48871.1 KT588080 AOE47990.1 LC017788 BAR64802.1 KX655904 AOG12853.1 ODYU01004533 SOQ44519.1 HM562971 ADR64682.1 KX585405 ARO70298.1 KY225536 ARO70548.1 KX084511 ARO76466.1 KQ458793 KPJ04976.1 KQ459984 KPJ19118.1 KU702634 AMM70661.1 RSAL01000162 RVE45396.1 KX778609 AON96599.1 AGBW02012180 OWR45511.1 PYGN01001739 PSN33061.1 FX982937 BAU20263.1 KK852704 KDR18055.1 KQ971372 EFA10559.2 ABLF02025841 GECU01030838 GECU01018545 JAS76868.1 JAS89161.1 GGOB01000056 MCH29452.1 ACPB03022988 ACPB03022989 DS235327 EEB14716.1 KK854187 PTY12807.1 KK107020 EZA62565.1 QOIP01000006 RLU21499.1 KQ981820 KYN35238.1 GBHO01031390 JAG12214.1 KQ435833 KOX71615.1 KQ763450 OAD55050.1 GL887596 EGI70677.1 KQ982254 KYQ58688.1 KQ979002 KYN26674.1 KZ288219 PBC32228.1 KQ977827 KYM99452.1 KQ976424 KYM88215.1 KQ415041 KOC58902.1 KQ435048 KZC14210.1 CH477324 EAT43539.1 UFQT01000650 SSX26109.1 GL435626 EFN73019.1 KB466333 RLZ02244.1 KX609448 ARN17855.1 JXUM01102368 KQ564722 KXJ71842.1 DS231840 EDS34587.1 AXCM01001403 NNAY01000077 OXU31196.1 AXCN02002267 CVRI01000057 CRL02414.1 CP012525 ALC43688.1 JRES01000714 KNC29001.1 AAAB01008964 EAA12883.4 CM002912 KMY98967.1 CH963847 EDW72727.2 JXJN01019455 CH902618 EDV39321.2 CCAG010008760 CH933809 EDW19302.1 KMY98968.1 CH954178 EDV51395.2 CM000159 EDW94108.1 AE014296 AAF50075.2 CH916366 EDV97099.1 OUUW01000002 SPP77051.1 AHN58056.1 CM000363 EDX10063.1 CH379069 EAL29662.2 CH479199 EDW29447.1 CH480815 EDW41076.1 ATLV01014965 KE524999 KFB39693.1 AJVK01007783 CH940647 EDW70920.1 LBMM01006286 KMQ90732.1
JAI18095.1 NWSH01003171 PCG66830.1 MG546654 AYD42277.1 JTDY01003244 KOB69888.1 MG820700 AXF48871.1 KT588080 AOE47990.1 LC017788 BAR64802.1 KX655904 AOG12853.1 ODYU01004533 SOQ44519.1 HM562971 ADR64682.1 KX585405 ARO70298.1 KY225536 ARO70548.1 KX084511 ARO76466.1 KQ458793 KPJ04976.1 KQ459984 KPJ19118.1 KU702634 AMM70661.1 RSAL01000162 RVE45396.1 KX778609 AON96599.1 AGBW02012180 OWR45511.1 PYGN01001739 PSN33061.1 FX982937 BAU20263.1 KK852704 KDR18055.1 KQ971372 EFA10559.2 ABLF02025841 GECU01030838 GECU01018545 JAS76868.1 JAS89161.1 GGOB01000056 MCH29452.1 ACPB03022988 ACPB03022989 DS235327 EEB14716.1 KK854187 PTY12807.1 KK107020 EZA62565.1 QOIP01000006 RLU21499.1 KQ981820 KYN35238.1 GBHO01031390 JAG12214.1 KQ435833 KOX71615.1 KQ763450 OAD55050.1 GL887596 EGI70677.1 KQ982254 KYQ58688.1 KQ979002 KYN26674.1 KZ288219 PBC32228.1 KQ977827 KYM99452.1 KQ976424 KYM88215.1 KQ415041 KOC58902.1 KQ435048 KZC14210.1 CH477324 EAT43539.1 UFQT01000650 SSX26109.1 GL435626 EFN73019.1 KB466333 RLZ02244.1 KX609448 ARN17855.1 JXUM01102368 KQ564722 KXJ71842.1 DS231840 EDS34587.1 AXCM01001403 NNAY01000077 OXU31196.1 AXCN02002267 CVRI01000057 CRL02414.1 CP012525 ALC43688.1 JRES01000714 KNC29001.1 AAAB01008964 EAA12883.4 CM002912 KMY98967.1 CH963847 EDW72727.2 JXJN01019455 CH902618 EDV39321.2 CCAG010008760 CH933809 EDW19302.1 KMY98968.1 CH954178 EDV51395.2 CM000159 EDW94108.1 AE014296 AAF50075.2 CH916366 EDV97099.1 OUUW01000002 SPP77051.1 AHN58056.1 CM000363 EDX10063.1 CH379069 EAL29662.2 CH479199 EDW29447.1 CH480815 EDW41076.1 ATLV01014965 KE524999 KFB39693.1 AJVK01007783 CH940647 EDW70920.1 LBMM01006286 KMQ90732.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000283053
+ More
UP000007151 UP000245037 UP000027135 UP000007266 UP000007819 UP000192223 UP000015103 UP000009046 UP000053097 UP000279307 UP000078541 UP000053105 UP000007755 UP000075809 UP000078492 UP000242457 UP000005203 UP000078542 UP000078540 UP000053825 UP000076502 UP000008820 UP000000311 UP000069940 UP000249989 UP000002320 UP000075920 UP000075883 UP000215335 UP000075886 UP000183832 UP000076408 UP000002358 UP000075900 UP000092553 UP000078200 UP000037069 UP000007062 UP000192221 UP000095301 UP000007798 UP000092443 UP000092460 UP000007801 UP000092444 UP000009192 UP000079169 UP000095300 UP000008711 UP000092445 UP000002282 UP000000803 UP000001070 UP000268350 UP000000304 UP000001819 UP000008744 UP000091820 UP000069272 UP000001292 UP000030765 UP000092462 UP000008792 UP000036403
UP000007151 UP000245037 UP000027135 UP000007266 UP000007819 UP000192223 UP000015103 UP000009046 UP000053097 UP000279307 UP000078541 UP000053105 UP000007755 UP000075809 UP000078492 UP000242457 UP000005203 UP000078542 UP000078540 UP000053825 UP000076502 UP000008820 UP000000311 UP000069940 UP000249989 UP000002320 UP000075920 UP000075883 UP000215335 UP000075886 UP000183832 UP000076408 UP000002358 UP000075900 UP000092553 UP000078200 UP000037069 UP000007062 UP000192221 UP000095301 UP000007798 UP000092443 UP000092460 UP000007801 UP000092444 UP000009192 UP000079169 UP000095300 UP000008711 UP000092445 UP000002282 UP000000803 UP000001070 UP000268350 UP000000304 UP000001819 UP000008744 UP000091820 UP000069272 UP000001292 UP000030765 UP000092462 UP000008792 UP000036403
Interpro
ProteinModelPortal
H9JAW2
A0A075TD94
A0A0K8TVJ6
A0A2A4J5W4
A0A386H8Z8
A0A0L7L303
+ More
A0A345BF47 A0A1B3B704 A0A0F7QK03 A0A1B3P5I4 A0A2H1VUJ7 E5FIA7 A0A2K8GKZ5 A0A2K8GL85 A0A1Y9TJT5 A0A194QNB4 A0A194RMR1 A0A140G9I2 A0A3S2LVR8 A0A1C9EGI0 A0A212EVL8 A0A2P8XM32 A0A0U5ARX4 A0A067RDK9 D6X0V7 J9KAI3 A0A1B6IQG3 A0A310SCX2 A0A1W4XI02 T1H8I6 E0VMW0 A0A2R7VZ94 A0A026X2P0 A0A3L8DNP4 A0A195F448 A0A0A9WWV2 A0A0M8ZUV9 A0A310S941 F4W4Y1 A0A151XEJ6 A0A151JLH6 A0A2A3EKI7 A0A087ZVT3 A0A195CF85 A0A195BNX6 A0A0L7QJU4 A0A154PSX2 Q17BA7 A0A336MB17 E2A0W6 A0A3L9LSY2 A0A1S4F9D5 A0A1W6L170 A0A182H107 B0W521 A0A182W7U8 A0A182MDX7 A0A232FKC0 A0A182Q3D6 A0A1J1ITH8 A0A182YGS4 K7IQ73 A0A182RVK5 A0A0M4EII4 A0A1A9VXX5 A0A0L0C9S2 A0A1S4GWS3 Q7Q3M6 A0A1W4W5W6 A0A1I8NEI8 A0A0J9RTM5 B4MKM1 A0A1A9XRY2 A0A1B0BS36 B3M3Y7 A0A1B0FPF1 B4L1C0 A0A0J9RTA8 A0A3Q0IMF8 A0A1I8PRW8 B3NGT8 A0A1A9ZKB3 B4PF26 Q9VTH3 B4J2L4 A0A3B0JWG1 X2JCB2 B4QPS2 Q2LZ47 B4GZM0 A0A1A9WUV8 A0A2K6VB67 A0A182F4J2 B4HEC2 A0A084VNZ9 A0A1B0EYB4 B4LCM8 A0A0J7KKB3
A0A345BF47 A0A1B3B704 A0A0F7QK03 A0A1B3P5I4 A0A2H1VUJ7 E5FIA7 A0A2K8GKZ5 A0A2K8GL85 A0A1Y9TJT5 A0A194QNB4 A0A194RMR1 A0A140G9I2 A0A3S2LVR8 A0A1C9EGI0 A0A212EVL8 A0A2P8XM32 A0A0U5ARX4 A0A067RDK9 D6X0V7 J9KAI3 A0A1B6IQG3 A0A310SCX2 A0A1W4XI02 T1H8I6 E0VMW0 A0A2R7VZ94 A0A026X2P0 A0A3L8DNP4 A0A195F448 A0A0A9WWV2 A0A0M8ZUV9 A0A310S941 F4W4Y1 A0A151XEJ6 A0A151JLH6 A0A2A3EKI7 A0A087ZVT3 A0A195CF85 A0A195BNX6 A0A0L7QJU4 A0A154PSX2 Q17BA7 A0A336MB17 E2A0W6 A0A3L9LSY2 A0A1S4F9D5 A0A1W6L170 A0A182H107 B0W521 A0A182W7U8 A0A182MDX7 A0A232FKC0 A0A182Q3D6 A0A1J1ITH8 A0A182YGS4 K7IQ73 A0A182RVK5 A0A0M4EII4 A0A1A9VXX5 A0A0L0C9S2 A0A1S4GWS3 Q7Q3M6 A0A1W4W5W6 A0A1I8NEI8 A0A0J9RTM5 B4MKM1 A0A1A9XRY2 A0A1B0BS36 B3M3Y7 A0A1B0FPF1 B4L1C0 A0A0J9RTA8 A0A3Q0IMF8 A0A1I8PRW8 B3NGT8 A0A1A9ZKB3 B4PF26 Q9VTH3 B4J2L4 A0A3B0JWG1 X2JCB2 B4QPS2 Q2LZ47 B4GZM0 A0A1A9WUV8 A0A2K6VB67 A0A182F4J2 B4HEC2 A0A084VNZ9 A0A1B0EYB4 B4LCM8 A0A0J7KKB3
PDB
4UQQ
E-value=0.0021569,
Score=95
Ontologies
GO
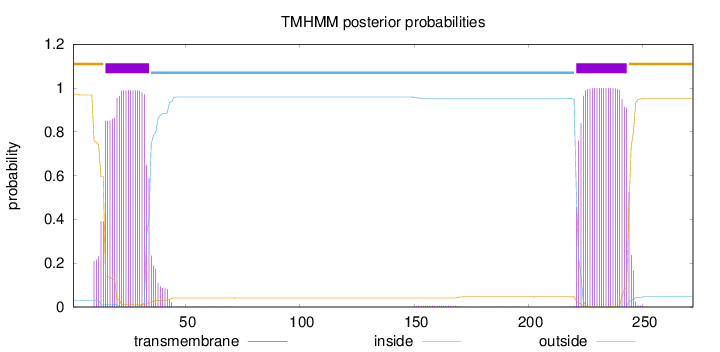
Topology
Length:
272
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.6787
Exp number, first 60 AAs:
20.8336
Total prob of N-in:
0.03121
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 34
inside
35 - 220
TMhelix
221 - 243
outside
244 - 272
Population Genetic Test Statistics
Pi
135.784896
Theta
152.51099
Tajima's D
-0.56367
CLR
0
CSRT
0.231738413079346
Interpretation
Uncertain
