Gene
KWMTBOMO05416
Pre Gene Modal
BGIBMGA006681
Annotation
Serine/threonine-protein_kinase_6_[Danaus_plexippus]
Full name
Aurora kinase A-A
+ More
Aurora kinase A-B
Aurora kinase A
Aurora kinase A-B
Aurora kinase A
Alternative Name
Aurora/IPL1-related kinase 1
Serine/threonine-protein kinase 6-A
Serine/threonine-protein kinase Eg2-A
Serine/threonine-protein kinase aurora-A
p46Eg265
Serine/threonine-protein kinase 6-B
Serine/threonine-protein kinase Eg2-B
p46XlEg22
Aurora 2
Aurora family kinase 1
Ipl1- and aurora-related kinase 1
Serine/threonine-protein kinase 6
Serine/threonine-protein kinase Ayk1
Serine/threonine-protein kinase 6-A
Serine/threonine-protein kinase Eg2-A
Serine/threonine-protein kinase aurora-A
p46Eg265
Serine/threonine-protein kinase 6-B
Serine/threonine-protein kinase Eg2-B
p46XlEg22
Aurora 2
Aurora family kinase 1
Ipl1- and aurora-related kinase 1
Serine/threonine-protein kinase 6
Serine/threonine-protein kinase Ayk1
Location in the cell
Cytoplasmic Reliability : 2.238
Sequence
CDS
ATGCCTACGGACAGTTTCAAAATCCCAGCCGTACCGTCTTCTCGGCCTTTATGTTCAAAAGCAAACGCCAGTGACAACAAAGACCAGACTTCTTCAAGACTTGAATTGAAACGTGACGATAAAAAGAAGACTTGGTCACTATCGGACTTCGATCTAGGACGACCGCTCGGTAAAGGGAAATTCGGCAATGTGTACTTAGCCCGTGAAAAAGAATCTCACTATGTCGTCGCATTGAAGGTTCTTTTTAAAAGCCAAATCTTGGATTCAGAAATAGAGCACCAAGTACGCCGTGAAGTTGAAATCCAGTGTCGACTACGTCATCCGAACATCTTACGCATGTATGGATACTTTCACGATGAAAAACGAATTTATTTAATATTAGAATACGCGAAACATGGCGCCCTTTACAAATTACTGAAGGAACGCGGCCGATTTGATGAAAAAACCGCTGCAATTTACATTCGTGATTTGACGAAGGCCCTTATTTATTGTCATACCAAGAAAGTTATACATCGTGATATAAAACCGGAAAATTTATTGATTGGTCACAACTGGGAACTTAAAATTGCTGATTTTGGCTGGTCCGTCCACTCGCCCTCATCAAGACGGATGACATTGTGCGGAACATTAGATTACCTGTCACCAGAAATGATTGAAGGAAAACCCCATAATTATGCAGTAGATATTTGGAGTTTAGGGGTTTTGTGTTACGAACTGCTTGTTGGATTGCCACCATTTGATGCCAAGGACTCTCATCAAACCTACAGAAAAATTAGATATGTCATAATTAAATATCCCGAATATATTTCTGAAAAAGCCAAAGATTTAATGGGAAAACTTTTAGTTATCGAGCCTGAAGAGAGGTTGCCACTATCAAATGTCTTAAAACACCCGTGGATTATAGAGAATGCTCCTGAAGGTGCTCATCCACCATTAATGAATGCTGAGCAAAAATAA
Protein
MPTDSFKIPAVPSSRPLCSKANASDNKDQTSSRLELKRDDKKKTWSLSDFDLGRPLGKGKFGNVYLAREKESHYVVALKVLFKSQILDSEIEHQVRREVEIQCRLRHPNILRMYGYFHDEKRIYLILEYAKHGALYKLLKERGRFDEKTAAIYIRDLTKALIYCHTKKVIHRDIKPENLLIGHNWELKIADFGWSVHSPSSRRMTLCGTLDYLSPEMIEGKPHNYAVDIWSLGVLCYELLVGLPPFDAKDSHQTYRKIRYVIIKYPEYISEKAKDLMGKLLVIEPEERLPLSNVLKHPWIIENAPEGAHPPLMNAEQK
Summary
Description
Mitotic serine/threonine kinases that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Phosphorylates numerous target proteins. Important for microtubule formation and/or stabilization.
Mitotic serine/threonine kinases that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Phosphorylates numerous target proteins. Important for microtubule formation and/or stabilization (By similarity).
Mitotic serine/threonine kinase that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Required for normal spindle positioning during mitosis and for the localization of NUMA1 and DCTN1 to the cell cortex during metaphase (By similarity). Required for initial activation of CDK1 at centrosomes. Phosphorylates numerous target proteins, including ARHGEF2, BORA, BRCA1, CDC25B, DLGP5, HDAC6, KIF2A, LATS2, NDEL1, PARD3, PPP1R2, PLK1, RASSF1, TACC3, p53/TP53 and TPX2. Regulates KIF2A tubulin depolymerase activity. Required for normal axon formation. Plays a role in microtubule remodeling during neurite extension. Important for microtubule formation and/or stabilization. Also acts as a key regulatory component of the p53/TP53 pathway, and particularly the checkpoint-response pathways critical for oncogenic transformation of cells, by phosphorylating and stabilizing p53/TP53. Phosphorylates its own inhibitors, the protein phosphatase type 1 (PP1) isoforms, to inhibit their activity. Necessary for proper cilia disassembly prior to mitosis.
Mitotic serine/threonine kinase that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Required for normal spindle positioning during mitosis and for the localization of NUMA1 and DCTN1 to the cell cortex during metaphase (By similarity). Required for initial activation of CDK1 at centrosomes. Phosphorylates numerous target proteins, including ARHGEF2, BORA, BRCA1, CDC25B, DLGP5, HDAC6, KIF2A, LATS2, NDEL1, PARD3, PPP1R2, PLK1, RASSF1, TACC3, p53/TP53 and TPX2. Regulates KIF2A tubulin depolymerase activity. Required for normal axon formation. Plays a role in microtubule remodeling during neurite extension. Important for microtubule formation and/or stabilization. Also acts as a key regulatory component of the p53/TP53 pathway, and particularly the checkpoint-response pathways critical for oncogenic transformation of cells, by phosphorylating and stabilizating p53/TP53. Phosphorylates its own inhibitors, the protein phosphatase type 1 (PP1) isoforms, to inhibit their activity (By similarity). Necessary for proper cilia disassembly prior to mitosis (By similarity). Interacts with SIRT2 (By similarity).
Mitotic serine/threonine kinases that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Phosphorylates numerous target proteins. Important for microtubule formation and/or stabilization (By similarity).
Mitotic serine/threonine kinase that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Required for normal spindle positioning during mitosis and for the localization of NUMA1 and DCTN1 to the cell cortex during metaphase (By similarity). Required for initial activation of CDK1 at centrosomes. Phosphorylates numerous target proteins, including ARHGEF2, BORA, BRCA1, CDC25B, DLGP5, HDAC6, KIF2A, LATS2, NDEL1, PARD3, PPP1R2, PLK1, RASSF1, TACC3, p53/TP53 and TPX2. Regulates KIF2A tubulin depolymerase activity. Required for normal axon formation. Plays a role in microtubule remodeling during neurite extension. Important for microtubule formation and/or stabilization. Also acts as a key regulatory component of the p53/TP53 pathway, and particularly the checkpoint-response pathways critical for oncogenic transformation of cells, by phosphorylating and stabilizing p53/TP53. Phosphorylates its own inhibitors, the protein phosphatase type 1 (PP1) isoforms, to inhibit their activity. Necessary for proper cilia disassembly prior to mitosis.
Mitotic serine/threonine kinase that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Required for normal spindle positioning during mitosis and for the localization of NUMA1 and DCTN1 to the cell cortex during metaphase (By similarity). Required for initial activation of CDK1 at centrosomes. Phosphorylates numerous target proteins, including ARHGEF2, BORA, BRCA1, CDC25B, DLGP5, HDAC6, KIF2A, LATS2, NDEL1, PARD3, PPP1R2, PLK1, RASSF1, TACC3, p53/TP53 and TPX2. Regulates KIF2A tubulin depolymerase activity. Required for normal axon formation. Plays a role in microtubule remodeling during neurite extension. Important for microtubule formation and/or stabilization. Also acts as a key regulatory component of the p53/TP53 pathway, and particularly the checkpoint-response pathways critical for oncogenic transformation of cells, by phosphorylating and stabilizating p53/TP53. Phosphorylates its own inhibitors, the protein phosphatase type 1 (PP1) isoforms, to inhibit their activity (By similarity). Necessary for proper cilia disassembly prior to mitosis (By similarity). Interacts with SIRT2 (By similarity).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with kif2c and kif11.
Interacts with CPEB1, JTB, TACC1, TPX2, PPP2CA, as well as with the protein phosphatase type 1 (PP1) isoforms PPP1CA, PPP1CB and PPP1CC (By similarity). Interacts also with its substrates ARHGEF2, BORA, BRCA1, KIF2A, PARD3, and p53/TP53. Interaction with BORA promotes phosphorylation of PLK1. Interacts with GADD45A, competing with its oligomerization (By similarity). Interacts with FBXL7 and PIFO. Interacts (via C-terminus) with AUNIP (via C-terminus) (By similarity). Identified in a complex with AUNIP and NIN (By similarity). Interacts with SIRT2 (By similarity). Interacts with FRY; this interaction facilitates AURKA-mediated PLK1 phosphorylation. Interacts with MYCN; interaction is phospho-independent and triggers AURKA activation; AURKA competes with FBXW7 for binding to unphosphorylated MYCN but not for binding to phosphorylated MYCN (By similarity). Interacts with HNRNPU (By similarity). Interacts with AAAS (By similarity).
Interacts with CPEB1, JTB, TACC1, TPX2, PPP2CA, as well as with the protein phosphatase type 1 (PP1) isoforms PPP1CA, PPP1CB and PPP1CC (By similarity). Interacts also with its substrates ARHGEF2, BORA, BRCA1, KIF2A, PARD3, and p53/TP53. Interaction with BORA promotes phosphorylation of PLK1. Interacts with FBXL7 and PIFO. Interacts with GADD45A, competing with its oligomerization (By similarity). Interacts (via C-terminus) with AUNIP (via C-terminus) (By similarity). Identified in a complex with AUNIP and NIN (By similarity). Interacts with SIRT2 (By similarity). Interacts with FRY; this interaction facilitates AURKA-mediated PLK1 phosphorylation (By similarity). Interacts with MYCN; interaction is phospho-independent and triggers AURKA activation; AURKA competes with FBXW7 for binding to unphosphorylated MYCN but not for binding to phosphorylated MYCN (By similarity). Interacts with HNRNPU (By similarity). Interacts with AAAS (By similarity).
Interacts with CPEB1, JTB, TACC1, TPX2, PPP2CA, as well as with the protein phosphatase type 1 (PP1) isoforms PPP1CA, PPP1CB and PPP1CC (By similarity). Interacts also with its substrates ARHGEF2, BORA, BRCA1, KIF2A, PARD3, and p53/TP53. Interaction with BORA promotes phosphorylation of PLK1. Interacts with GADD45A, competing with its oligomerization (By similarity). Interacts with FBXL7 and PIFO. Interacts (via C-terminus) with AUNIP (via C-terminus) (By similarity). Identified in a complex with AUNIP and NIN (By similarity). Interacts with SIRT2 (By similarity). Interacts with FRY; this interaction facilitates AURKA-mediated PLK1 phosphorylation. Interacts with MYCN; interaction is phospho-independent and triggers AURKA activation; AURKA competes with FBXW7 for binding to unphosphorylated MYCN but not for binding to phosphorylated MYCN (By similarity). Interacts with HNRNPU (By similarity). Interacts with AAAS (By similarity).
Interacts with CPEB1, JTB, TACC1, TPX2, PPP2CA, as well as with the protein phosphatase type 1 (PP1) isoforms PPP1CA, PPP1CB and PPP1CC (By similarity). Interacts also with its substrates ARHGEF2, BORA, BRCA1, KIF2A, PARD3, and p53/TP53. Interaction with BORA promotes phosphorylation of PLK1. Interacts with FBXL7 and PIFO. Interacts with GADD45A, competing with its oligomerization (By similarity). Interacts (via C-terminus) with AUNIP (via C-terminus) (By similarity). Identified in a complex with AUNIP and NIN (By similarity). Interacts with SIRT2 (By similarity). Interacts with FRY; this interaction facilitates AURKA-mediated PLK1 phosphorylation (By similarity). Interacts with MYCN; interaction is phospho-independent and triggers AURKA activation; AURKA competes with FBXW7 for binding to unphosphorylated MYCN but not for binding to phosphorylated MYCN (By similarity). Interacts with HNRNPU (By similarity). Interacts with AAAS (By similarity).
Miscellaneous
Centrosome amplification can occur when the cycles are uncoupled, and this amplification is associated with cancer and with an increase in the levels of chromosomal instability.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. Aurora subfamily.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. Aurora subfamily.
Keywords
ATP-binding
Cell cycle
Cell division
Cytoplasm
Cytoskeleton
Kinase
Microtubule
Mitosis
Nucleotide-binding
Phosphoprotein
Transferase
3D-structure
Alternative splicing
Cell projection
Cilium
Complete proteome
Isopeptide bond
Proto-oncogene
Reference proteome
Ubl conjugation
Feature
chain Aurora kinase A-A
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JAY6
A0A2A4J4B0
A0A2H1VUQ0
A0A212EVI6
A0A0L7L337
S4PLF3
+ More
A0A1C9EGH0 A0A194QJ70 A0A194RNY2 M3XKI8 A0A1L8ELH7 A0A1L8ESU8 Q91820 Q91819 A0A2A3EAH8 A0A087ZXW3 G3HBL6 Q28HZ5 B5DFP5 A0A091V3E5 A0A087VKZ9 A0A1U7R6R7 A0A091H823 Q3MHU2 A0A151NYV8 Q7TNK2 Q3TEY6 P97477 K7G824 A0A2T7NIA0 P59241 A0A2P4Y0B0 A0A099YWA5 P97477-2 Q010G9 G3WJ52 A0A1X7VED6 A0A225WSH3 D0NUI6 F7ASA8 V8NVD6 H3B947 A0A2I0TNT3 A0A2I0M5W6 A0A1W4WSM8 V4A8Z0 L1IU42 A0A232EXQ7 K7JAY3 A0A1U7QWD0 A0A091W399 H9GMT8 A0A093HWJ7 E1BVE1 A0A060Y833 G1PAC6 R0KDE3 U3IAE4 F6S2E1 W2W8U2 W2IBC2 W2KG09 V9EGC6 A0A0W8CBA6 W2PNT8 Q8BP87 W2YJF0 A0A080ZGT4 W2MNH0 H0WPW8 A0A1J1IC17 A0A2Y9IM04 D7ELW1 A0A2Y9Q6F5 A0A2P4T2Q2 A0A1W2W3M3 A0A0W8D4G9 A0A1S3FT78 A0A1A6FYP0 A0A2Y9QCE6 A0A341BTD3 A0A1S3WNV3 A0A2P4XCZ7 A0A087QS09 A0A226PHT5 C1MQG5 A0A3Q1GSL9 A0A1V4KE16 A0A154P152 A0A2Y9ELJ1 F7A0P0 A4S3J2 A0A226MHJ8 A0A2Y9INE5 A0A3B3RXY0 A0A3P8SMK4 A0A3P8SMT4 A0A2Y9ELJ6 B5X2V6 I6V4W2
A0A1C9EGH0 A0A194QJ70 A0A194RNY2 M3XKI8 A0A1L8ELH7 A0A1L8ESU8 Q91820 Q91819 A0A2A3EAH8 A0A087ZXW3 G3HBL6 Q28HZ5 B5DFP5 A0A091V3E5 A0A087VKZ9 A0A1U7R6R7 A0A091H823 Q3MHU2 A0A151NYV8 Q7TNK2 Q3TEY6 P97477 K7G824 A0A2T7NIA0 P59241 A0A2P4Y0B0 A0A099YWA5 P97477-2 Q010G9 G3WJ52 A0A1X7VED6 A0A225WSH3 D0NUI6 F7ASA8 V8NVD6 H3B947 A0A2I0TNT3 A0A2I0M5W6 A0A1W4WSM8 V4A8Z0 L1IU42 A0A232EXQ7 K7JAY3 A0A1U7QWD0 A0A091W399 H9GMT8 A0A093HWJ7 E1BVE1 A0A060Y833 G1PAC6 R0KDE3 U3IAE4 F6S2E1 W2W8U2 W2IBC2 W2KG09 V9EGC6 A0A0W8CBA6 W2PNT8 Q8BP87 W2YJF0 A0A080ZGT4 W2MNH0 H0WPW8 A0A1J1IC17 A0A2Y9IM04 D7ELW1 A0A2Y9Q6F5 A0A2P4T2Q2 A0A1W2W3M3 A0A0W8D4G9 A0A1S3FT78 A0A1A6FYP0 A0A2Y9QCE6 A0A341BTD3 A0A1S3WNV3 A0A2P4XCZ7 A0A087QS09 A0A226PHT5 C1MQG5 A0A3Q1GSL9 A0A1V4KE16 A0A154P152 A0A2Y9ELJ1 F7A0P0 A4S3J2 A0A226MHJ8 A0A2Y9INE5 A0A3B3RXY0 A0A3P8SMK4 A0A3P8SMT4 A0A2Y9ELJ6 B5X2V6 I6V4W2
EC Number
2.7.11.1
Pubmed
19121390
22118469
26227816
23622113
26354079
9215903
+ More
27762356 9454730 10329703 18434591 21804562 29704459 20431018 15489334 15057822 15632090 22293439 10349636 11042159 11076861 11217851 12466851 12040188 16141073 9245792 9205101 9514916 16141072 15793587 19075002 19668197 21183079 20643351 22306998 22753416 18579375 18678489 23807208 17381049 12124350 19812038 28186564 16868079 21709235 19741609 17495919 24297900 23371554 23254933 23201678 28648823 20075255 21881562 15592404 24755649 21993624 23749191 18464734 18362917 19820115 24621616 19359590 12481130 15114417 17460045 29240929 20433749
27762356 9454730 10329703 18434591 21804562 29704459 20431018 15489334 15057822 15632090 22293439 10349636 11042159 11076861 11217851 12466851 12040188 16141073 9245792 9205101 9514916 16141072 15793587 19075002 19668197 21183079 20643351 22306998 22753416 18579375 18678489 23807208 17381049 12124350 19812038 28186564 16868079 21709235 19741609 17495919 24297900 23371554 23254933 23201678 28648823 20075255 21881562 15592404 24755649 21993624 23749191 18464734 18362917 19820115 24621616 19359590 12481130 15114417 17460045 29240929 20433749
EMBL
BABH01008325
NWSH01003171
PCG66831.1
ODYU01004533
SOQ44518.1
AGBW02012180
+ More
OWR45510.1 JTDY01003244 KOB69887.1 GAIX01004055 JAA88505.1 KX778609 AON96600.1 KQ458793 KPJ04975.1 KQ459984 KPJ19119.1 AFYH01027207 AFYH01027208 AFYH01027209 CM004483 OCT60187.1 CM004482 OCT62407.1 Z17207 BC072133 Z17206 BC075177 KZ288308 PBC28700.1 JH000271 RAZU01000236 EGW11017.1 RLQ63681.1 AAMC01123675 AAMC01123676 CR760668 KV460429 CAJ82185.1 OCA18246.1 BC169141 AAI69141.1 KL410577 KFQ97303.1 KL497174 KFO13291.1 KL525231 KFO90967.1 AC131024 BC104676 CH474062 AAI04677.1 EDL85147.1 AKHW03001485 KYO42062.1 AY336976 AAQ16152.1 AK145968 AK169363 CH466551 BAE26793.1 BAE41112.1 EDL06601.1 U80932 AF007817 U69106 AK085861 BC005425 BC014711 AGCU01129861 AGCU01129862 AGCU01129863 PZQS01000012 PVD20894.1 AF537333 NCKW01006561 POM71234.1 KL885837 KGL73233.1 CAID01000010 CAL56063.1 AEFK01024103 NBNE01000309 OWZ20585.1 DS028164 EEY65332.1 AZIM01001870 ETE65492.1 KZ508257 PKU35440.1 AKCR02000035 PKK25077.1 KB202544 ESO89771.1 JH993038 EKX39637.1 NNAY01001682 OXU23236.1 AAZX01011334 KK734682 KFR10122.1 AAWZ02020085 KL206811 KFV86021.1 AADN05000479 FR908224 CDQ87896.1 AAPE02030398 KB742479 EOB08007.1 ADON01010430 ANIX01003357 ETP06962.1 KI688439 KI675217 ETK77321.1 ETL30767.1 KI681915 ETL84012.1 ANIZ01002956 ETI37112.1 LNFO01004195 KUF81362.1 KI669616 ETN02658.1 AK077513 BAC36838.1 ANIY01003537 ETP35051.1 ANJA01003141 ETO65845.1 KI695163 ETM37198.1 AAQR03050642 AAQR03050643 AAQR03050644 CVRI01000047 CRK97821.1 DS497934 EFA12426.1 PPHD01011082 POI30636.1 LNFO01001495 KUF91020.1 LZPO01113206 OBS58680.1 NCKW01011496 POM63420.1 KL225851 KFM04013.1 AWGT02000072 OXB79254.1 GG663738 EEH57705.1 LSYS01003456 OPJ82719.1 KQ434796 KZC05563.1 EAAA01002391 CP000590 ABO98397.1 MCFN01000860 OXB54786.1 BT045375 ACI33637.1 JX020948 AFN06392.1
OWR45510.1 JTDY01003244 KOB69887.1 GAIX01004055 JAA88505.1 KX778609 AON96600.1 KQ458793 KPJ04975.1 KQ459984 KPJ19119.1 AFYH01027207 AFYH01027208 AFYH01027209 CM004483 OCT60187.1 CM004482 OCT62407.1 Z17207 BC072133 Z17206 BC075177 KZ288308 PBC28700.1 JH000271 RAZU01000236 EGW11017.1 RLQ63681.1 AAMC01123675 AAMC01123676 CR760668 KV460429 CAJ82185.1 OCA18246.1 BC169141 AAI69141.1 KL410577 KFQ97303.1 KL497174 KFO13291.1 KL525231 KFO90967.1 AC131024 BC104676 CH474062 AAI04677.1 EDL85147.1 AKHW03001485 KYO42062.1 AY336976 AAQ16152.1 AK145968 AK169363 CH466551 BAE26793.1 BAE41112.1 EDL06601.1 U80932 AF007817 U69106 AK085861 BC005425 BC014711 AGCU01129861 AGCU01129862 AGCU01129863 PZQS01000012 PVD20894.1 AF537333 NCKW01006561 POM71234.1 KL885837 KGL73233.1 CAID01000010 CAL56063.1 AEFK01024103 NBNE01000309 OWZ20585.1 DS028164 EEY65332.1 AZIM01001870 ETE65492.1 KZ508257 PKU35440.1 AKCR02000035 PKK25077.1 KB202544 ESO89771.1 JH993038 EKX39637.1 NNAY01001682 OXU23236.1 AAZX01011334 KK734682 KFR10122.1 AAWZ02020085 KL206811 KFV86021.1 AADN05000479 FR908224 CDQ87896.1 AAPE02030398 KB742479 EOB08007.1 ADON01010430 ANIX01003357 ETP06962.1 KI688439 KI675217 ETK77321.1 ETL30767.1 KI681915 ETL84012.1 ANIZ01002956 ETI37112.1 LNFO01004195 KUF81362.1 KI669616 ETN02658.1 AK077513 BAC36838.1 ANIY01003537 ETP35051.1 ANJA01003141 ETO65845.1 KI695163 ETM37198.1 AAQR03050642 AAQR03050643 AAQR03050644 CVRI01000047 CRK97821.1 DS497934 EFA12426.1 PPHD01011082 POI30636.1 LNFO01001495 KUF91020.1 LZPO01113206 OBS58680.1 NCKW01011496 POM63420.1 KL225851 KFM04013.1 AWGT02000072 OXB79254.1 GG663738 EEH57705.1 LSYS01003456 OPJ82719.1 KQ434796 KZC05563.1 EAAA01002391 CP000590 ABO98397.1 MCFN01000860 OXB54786.1 BT045375 ACI33637.1 JX020948 AFN06392.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053268
UP000053240
+ More
UP000008672 UP000186698 UP000242457 UP000005203 UP000001075 UP000273346 UP000008143 UP000053283 UP000189705 UP000002494 UP000050525 UP000000589 UP000007267 UP000245119 UP000053641 UP000009170 UP000007648 UP000007879 UP000198211 UP000006643 UP000002280 UP000053872 UP000192223 UP000030746 UP000011087 UP000215335 UP000002358 UP000189706 UP000053605 UP000001646 UP000053584 UP000000539 UP000193380 UP000001074 UP000016666 UP000002279 UP000018958 UP000053236 UP000053864 UP000054423 UP000018721 UP000052943 UP000018817 UP000018948 UP000028582 UP000054532 UP000005225 UP000183832 UP000248482 UP000007266 UP000248483 UP000081671 UP000092124 UP000252040 UP000079721 UP000053286 UP000198419 UP000001876 UP000257200 UP000190648 UP000076502 UP000008144 UP000001568 UP000198323 UP000261540 UP000265080 UP000248484 UP000087266
UP000008672 UP000186698 UP000242457 UP000005203 UP000001075 UP000273346 UP000008143 UP000053283 UP000189705 UP000002494 UP000050525 UP000000589 UP000007267 UP000245119 UP000053641 UP000009170 UP000007648 UP000007879 UP000198211 UP000006643 UP000002280 UP000053872 UP000192223 UP000030746 UP000011087 UP000215335 UP000002358 UP000189706 UP000053605 UP000001646 UP000053584 UP000000539 UP000193380 UP000001074 UP000016666 UP000002279 UP000018958 UP000053236 UP000053864 UP000054423 UP000018721 UP000052943 UP000018817 UP000018948 UP000028582 UP000054532 UP000005225 UP000183832 UP000248482 UP000007266 UP000248483 UP000081671 UP000092124 UP000252040 UP000079721 UP000053286 UP000198419 UP000001876 UP000257200 UP000190648 UP000076502 UP000008144 UP000001568 UP000198323 UP000261540 UP000265080 UP000248484 UP000087266
Interpro
Gene 3D
ProteinModelPortal
H9JAY6
A0A2A4J4B0
A0A2H1VUQ0
A0A212EVI6
A0A0L7L337
S4PLF3
+ More
A0A1C9EGH0 A0A194QJ70 A0A194RNY2 M3XKI8 A0A1L8ELH7 A0A1L8ESU8 Q91820 Q91819 A0A2A3EAH8 A0A087ZXW3 G3HBL6 Q28HZ5 B5DFP5 A0A091V3E5 A0A087VKZ9 A0A1U7R6R7 A0A091H823 Q3MHU2 A0A151NYV8 Q7TNK2 Q3TEY6 P97477 K7G824 A0A2T7NIA0 P59241 A0A2P4Y0B0 A0A099YWA5 P97477-2 Q010G9 G3WJ52 A0A1X7VED6 A0A225WSH3 D0NUI6 F7ASA8 V8NVD6 H3B947 A0A2I0TNT3 A0A2I0M5W6 A0A1W4WSM8 V4A8Z0 L1IU42 A0A232EXQ7 K7JAY3 A0A1U7QWD0 A0A091W399 H9GMT8 A0A093HWJ7 E1BVE1 A0A060Y833 G1PAC6 R0KDE3 U3IAE4 F6S2E1 W2W8U2 W2IBC2 W2KG09 V9EGC6 A0A0W8CBA6 W2PNT8 Q8BP87 W2YJF0 A0A080ZGT4 W2MNH0 H0WPW8 A0A1J1IC17 A0A2Y9IM04 D7ELW1 A0A2Y9Q6F5 A0A2P4T2Q2 A0A1W2W3M3 A0A0W8D4G9 A0A1S3FT78 A0A1A6FYP0 A0A2Y9QCE6 A0A341BTD3 A0A1S3WNV3 A0A2P4XCZ7 A0A087QS09 A0A226PHT5 C1MQG5 A0A3Q1GSL9 A0A1V4KE16 A0A154P152 A0A2Y9ELJ1 F7A0P0 A4S3J2 A0A226MHJ8 A0A2Y9INE5 A0A3B3RXY0 A0A3P8SMK4 A0A3P8SMT4 A0A2Y9ELJ6 B5X2V6 I6V4W2
A0A1C9EGH0 A0A194QJ70 A0A194RNY2 M3XKI8 A0A1L8ELH7 A0A1L8ESU8 Q91820 Q91819 A0A2A3EAH8 A0A087ZXW3 G3HBL6 Q28HZ5 B5DFP5 A0A091V3E5 A0A087VKZ9 A0A1U7R6R7 A0A091H823 Q3MHU2 A0A151NYV8 Q7TNK2 Q3TEY6 P97477 K7G824 A0A2T7NIA0 P59241 A0A2P4Y0B0 A0A099YWA5 P97477-2 Q010G9 G3WJ52 A0A1X7VED6 A0A225WSH3 D0NUI6 F7ASA8 V8NVD6 H3B947 A0A2I0TNT3 A0A2I0M5W6 A0A1W4WSM8 V4A8Z0 L1IU42 A0A232EXQ7 K7JAY3 A0A1U7QWD0 A0A091W399 H9GMT8 A0A093HWJ7 E1BVE1 A0A060Y833 G1PAC6 R0KDE3 U3IAE4 F6S2E1 W2W8U2 W2IBC2 W2KG09 V9EGC6 A0A0W8CBA6 W2PNT8 Q8BP87 W2YJF0 A0A080ZGT4 W2MNH0 H0WPW8 A0A1J1IC17 A0A2Y9IM04 D7ELW1 A0A2Y9Q6F5 A0A2P4T2Q2 A0A1W2W3M3 A0A0W8D4G9 A0A1S3FT78 A0A1A6FYP0 A0A2Y9QCE6 A0A341BTD3 A0A1S3WNV3 A0A2P4XCZ7 A0A087QS09 A0A226PHT5 C1MQG5 A0A3Q1GSL9 A0A1V4KE16 A0A154P152 A0A2Y9ELJ1 F7A0P0 A4S3J2 A0A226MHJ8 A0A2Y9INE5 A0A3B3RXY0 A0A3P8SMK4 A0A3P8SMT4 A0A2Y9ELJ6 B5X2V6 I6V4W2
PDB
3DJ7
E-value=1.76054e-101,
Score=942
Ontologies
GO
GO:0005524
GO:0004674
GO:0004672
GO:0007100
GO:0007052
GO:0000212
GO:0090307
GO:0005737
GO:0005876
GO:0046777
GO:0000922
GO:0018105
GO:0019894
GO:0008017
GO:0051301
GO:0005813
GO:0000780
GO:0051233
GO:0035174
GO:0031616
GO:0032133
GO:0032465
GO:0007057
GO:1990138
GO:0031625
GO:0043203
GO:0072687
GO:0032091
GO:0009948
GO:0019901
GO:0005829
GO:0043066
GO:0045120
GO:0071539
GO:0032436
GO:0048471
GO:0051642
GO:0042585
GO:0046982
GO:0031647
GO:0097421
GO:1900195
GO:0097431
GO:0009611
GO:0005814
GO:0005634
GO:0000278
GO:0005815
GO:0007051
GO:0072686
GO:0000226
GO:0005654
GO:0005819
GO:0006468
GO:0015630
GO:0005929
GO:0051321
GO:0007098
GO:0032355
GO:0030030
GO:0006879
GO:0005623
GO:0008199
GO:0016567
GO:0006810
GO:0004889
GO:0005216
GO:0045211
GO:0016491
GO:0003824
PANTHER
Topology
Subcellular location
Cytoplasm
Cytoskeleton
Spindle
Microtubule organizing center
Centrosome
Spindle pole Localizes to the spindle pole during mitosis especially from prophase through anaphase. Partially colocalized with gamma tubulin in the centrosome, from S to M phase (By similarity). With evidence from 1 publications.
Cilium basal body Localizes on centrosomes in interphase cells and at each spindle pole in mitosis (PubMed:9245792). Associates with both the pericentriolar material (PCM) and centrioles (By similarity). Colocalized with SIRT2 at centrosome (By similarity). Detected at the neurite hillock in developing neurons (PubMed:19668197). The localization to the spindle poles is regulated by AAAS (By similarity). With evidence from 4 publications.
Centriole Localizes on centrosomes in interphase cells and at each spindle pole in mitosis (PubMed:9245792). Associates with both the pericentriolar material (PCM) and centrioles (By similarity). Colocalized with SIRT2 at centrosome (By similarity). Detected at the neurite hillock in developing neurons (PubMed:19668197). The localization to the spindle poles is regulated by AAAS (By similarity). With evidence from 4 publications.
Cytoskeleton
Spindle
Microtubule organizing center
Centrosome
Spindle pole Localizes to the spindle pole during mitosis especially from prophase through anaphase. Partially colocalized with gamma tubulin in the centrosome, from S to M phase (By similarity). With evidence from 1 publications.
Cilium basal body Localizes on centrosomes in interphase cells and at each spindle pole in mitosis (PubMed:9245792). Associates with both the pericentriolar material (PCM) and centrioles (By similarity). Colocalized with SIRT2 at centrosome (By similarity). Detected at the neurite hillock in developing neurons (PubMed:19668197). The localization to the spindle poles is regulated by AAAS (By similarity). With evidence from 4 publications.
Centriole Localizes on centrosomes in interphase cells and at each spindle pole in mitosis (PubMed:9245792). Associates with both the pericentriolar material (PCM) and centrioles (By similarity). Colocalized with SIRT2 at centrosome (By similarity). Detected at the neurite hillock in developing neurons (PubMed:19668197). The localization to the spindle poles is regulated by AAAS (By similarity). With evidence from 4 publications.
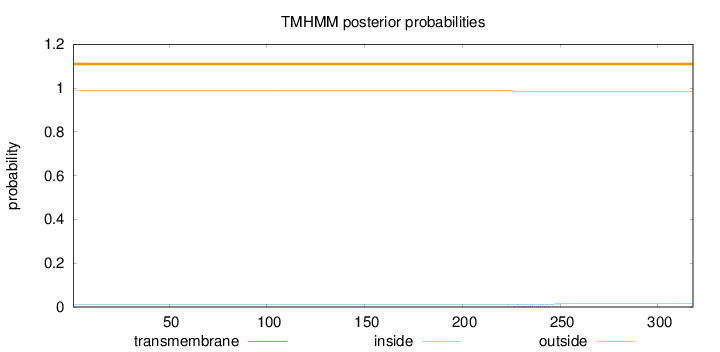
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08457
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01113
outside
1 - 318
Population Genetic Test Statistics
Pi
141.161235
Theta
179.237711
Tajima's D
-0.613503
CLR
1.223718
CSRT
0.215439228038598
Interpretation
Uncertain
