Gene
KWMTBOMO05411
Pre Gene Modal
BGIBMGA006680
Annotation
PREDICTED:_probable_DNA_replication_complex_GINS_protein_PSF2_[Amyelois_transitella]
Full name
DNA replication complex GINS protein PSF2
+ More
Probable DNA replication complex GINS protein PSF2
Probable DNA replication complex GINS protein PSF2
Alternative Name
GINS complex subunit 2
Location in the cell
Cytoplasmic Reliability : 1.451 Mitochondrial Reliability : 1.708
Sequence
CDS
ATGGAACCTTATGAAATCGAGTTTATTGGTGAGAATCGGATAATCAGTATAACACCAAACTTTACCCATGATAAAATATACTTGATATGCGGTGAATTTGGTCCGTTTCGTGCTGGTTTACCTGTGAACGTACCACTCTGGCTGGCTATCATGCTAAAGCAGAAACAAAAGTGCCATGTTATCCCTCCTGACTGGATGGACGTGGAAGTTTTGGAGAATATTAAAGAAGAGGAAAAGCGTTCCAGATTCTTCACAAAGATGCCAAATGAACACTATATGGTGGAAGCCAAACTGATACTTGGTTCAGCTGCCGAAGATGTTCCCAATGCACCAGAAATAAAGACTATTATCAAAGACATCTGGGATATAAGAATGTCTAAGTTACGAACATCAATGGATGCATTGATGAAAACTGGAGGTGGCTATGGCAGATTAGATCATTTAACAATGATGGAAATAAACTCAGTGAAACCTCTTCTACCTGCTGCTATGGACAACCTCTTAAGGATGCAAATGAAAACGAAAAAGACAGCACCATCGCTCAATTTAAATAGTTCAACCTTCAGTCAATCAGGGAGCTCTCAAAATACATAG
Protein
MEPYEIEFIGENRIISITPNFTHDKIYLICGEFGPFRAGLPVNVPLWLAIMLKQKQKCHVIPPDWMDVEVLENIKEEEKRSRFFTKMPNEHYMVEAKLILGSAAEDVPNAPEIKTIIKDIWDIRMSKLRTSMDALMKTGGGYGRLDHLTMMEINSVKPLLPAAMDNLLRMQMKTKKTAPSLNLNSSTFSQSGSSQNT
Summary
Description
The GINS complex plays an essential role in the initiation of DNA replication.
Subunit
Component of the GINS complex.
Component of the GINS complex which is a heterotetramer of Sld5, Psf1, Psf2 and Psf3.
Component of the GINS complex which is a heterotetramer of Sld5, Psf1, Psf2 and Psf3.
Similarity
Belongs to the GINS2/PSF2 family.
Keywords
Complete proteome
DNA replication
Nucleus
Reference proteome
Feature
chain Probable DNA replication complex GINS protein PSF2
Uniprot
H9JAY5
A0A1E1WP58
A0A1C9EGI7
A0A0L7KWA7
A0A2H1V7P2
A0A0N1INJ5
+ More
A0A194QNA9 A0A212EVH9 S4PLH1 A0A067QTG0 A0A2J7RRZ8 A0A2P8Y6T8 E9GHY1 A0A310SFE3 B0WD86 W8C889 A0A1B0A5Y2 A0A0N0BC54 A0A182GEW5 Q175H4 A0A0K8TKA9 D3TR45 A0A1A9V6W8 A0A1Q3EW89 A0A088A6B1 A0A2A3EAY0 A0A195DMV1 A0A1A9X614 A0A154PCQ7 A0A1A9WDL8 A0A1Y1M4S0 A0A195AWB3 A0A158P1W4 A0A3B0JC07 F4W642 A0A151I7X8 A0A0L7RJB2 E2AUX7 A0A162SE06 A0A195ERA8 A0A151WP32 A0A232FEH8 A0A0P6AGA1 A0A026WB65 E2B8N4 A0A1I8QEP2 A0A0K8W1V7 A0A182JKD0 A0A084VGK0 K7J3J2 A0A034WMY9 A0A1I8MQ31 A0A0A1X6Z9 A0A0L0CGI4 A0A182W8N7 T1G2N2 A0A182QKS7 J3JWM7 A0A182VCF8 A0A182X987 Q5TXV1 A0A182Y670 A0A182KZ66 A0A182HR63 A0A182TRK6 A0A2C9JNB1 A0A194ALK1 A0A182F350 A0A182LW41 A0A182RWN4 D6X0V8 B4MDS0 B5DIX7 B4GPW2 A0A1B6CNC0 B3N3R3 A0A1W4WAI5 B4JQE1 A0A182NNZ4 B4NYJ5 V3ZK44 W5JUU6 B3MU45 B4MZP3 B4QA63 B4I382 Q9VQY9 A0A2B4RX52 A0A1S4EMW1 A0A087SXK8 B5DIX8 A0A0J9QVI8 A0A2L2YP27 V5H5P6 B4KES6 A0A210PNE2 K1R5T3 A0A1S3HQ06 A0A0M5J8Z0 A0A182PXD8
A0A194QNA9 A0A212EVH9 S4PLH1 A0A067QTG0 A0A2J7RRZ8 A0A2P8Y6T8 E9GHY1 A0A310SFE3 B0WD86 W8C889 A0A1B0A5Y2 A0A0N0BC54 A0A182GEW5 Q175H4 A0A0K8TKA9 D3TR45 A0A1A9V6W8 A0A1Q3EW89 A0A088A6B1 A0A2A3EAY0 A0A195DMV1 A0A1A9X614 A0A154PCQ7 A0A1A9WDL8 A0A1Y1M4S0 A0A195AWB3 A0A158P1W4 A0A3B0JC07 F4W642 A0A151I7X8 A0A0L7RJB2 E2AUX7 A0A162SE06 A0A195ERA8 A0A151WP32 A0A232FEH8 A0A0P6AGA1 A0A026WB65 E2B8N4 A0A1I8QEP2 A0A0K8W1V7 A0A182JKD0 A0A084VGK0 K7J3J2 A0A034WMY9 A0A1I8MQ31 A0A0A1X6Z9 A0A0L0CGI4 A0A182W8N7 T1G2N2 A0A182QKS7 J3JWM7 A0A182VCF8 A0A182X987 Q5TXV1 A0A182Y670 A0A182KZ66 A0A182HR63 A0A182TRK6 A0A2C9JNB1 A0A194ALK1 A0A182F350 A0A182LW41 A0A182RWN4 D6X0V8 B4MDS0 B5DIX7 B4GPW2 A0A1B6CNC0 B3N3R3 A0A1W4WAI5 B4JQE1 A0A182NNZ4 B4NYJ5 V3ZK44 W5JUU6 B3MU45 B4MZP3 B4QA63 B4I382 Q9VQY9 A0A2B4RX52 A0A1S4EMW1 A0A087SXK8 B5DIX8 A0A0J9QVI8 A0A2L2YP27 V5H5P6 B4KES6 A0A210PNE2 K1R5T3 A0A1S3HQ06 A0A0M5J8Z0 A0A182PXD8
Pubmed
19121390
26227816
26354079
22118469
23622113
24845553
+ More
29403074 21292972 24495485 26483478 17510324 26369729 20353571 28004739 21347285 21719571 20798317 28648823 24508170 30249741 24438588 20075255 25348373 25315136 25830018 26108605 23254933 22516182 23537049 9087549 12364791 14747013 17210077 25244985 20966253 15562597 18362917 19820115 17994087 15632085 17550304 20920257 23761445 10731132 12537572 12537569 22936249 26561354 25765539 18057021 28812685 22992520
29403074 21292972 24495485 26483478 17510324 26369729 20353571 28004739 21347285 21719571 20798317 28648823 24508170 30249741 24438588 20075255 25348373 25315136 25830018 26108605 23254933 22516182 23537049 9087549 12364791 14747013 17210077 25244985 20966253 15562597 18362917 19820115 17994087 15632085 17550304 20920257 23761445 10731132 12537572 12537569 22936249 26561354 25765539 18057021 28812685 22992520
EMBL
BABH01008312
GDQN01002428
JAT88626.1
KX778609
AON96604.1
JTDY01005071
+ More
KOB67400.1 ODYU01001085 SOQ36827.1 KQ461033 KPJ09973.1 KQ458793 KPJ04971.1 AGBW02012180 OWR45505.1 GAIX01004030 JAA88530.1 KK853292 KDR08843.1 NEVH01000596 PNF43610.1 PYGN01000858 PSN39966.1 GL732545 EFX80914.1 KQ766110 OAD53736.1 DS231895 EDS44358.1 GAMC01000057 JAC06499.1 KQ435922 KOX68504.1 JXUM01010543 KQ560311 KXJ83110.1 CH477400 EAT41746.1 GDAI01002841 JAI14762.1 EZ423897 ADD20173.1 GFDL01015577 JAV19468.1 KZ288300 PBC28887.1 KQ980724 KYN14151.1 KQ434874 KZC09709.1 GEZM01040674 JAV80772.1 KQ976731 KYM76325.1 ADTU01006902 OUUW01000004 SPP79894.1 GL887707 EGI70177.1 KQ978397 KYM94208.1 KQ414582 KOC70914.1 GL442912 EFN62760.1 LRGB01000044 KZS21211.1 KQ982021 KYN30414.1 KQ982892 KYQ49624.1 NNAY01000390 OXU28717.1 GDIP01031145 JAM72570.1 KK107293 QOIP01000005 EZA53178.1 RLU22629.1 GL446361 EFN87940.1 GDHF01007499 JAI44815.1 ATLV01012963 KE524828 KFB37094.1 GAKP01002016 JAC56936.1 GBXI01007410 JAD06882.1 JRES01000438 KNC31322.1 AMQM01003587 KB096222 ESO07225.1 AXCN02002100 APGK01042893 BT127645 KB741009 KB632250 AEE62607.1 ENN75598.1 ERL90592.1 AAAB01006447 EAL42206.2 APCN01001653 GELH01000956 GELH01000955 JAS03317.1 AXCM01000708 KQ971372 EFA10560.2 CH940661 EDW71331.1 CH379060 EDY70270.1 CH479187 EDW39634.1 GEDC01030061 GEDC01022342 JAS07237.1 JAS14956.1 CH954177 EDV57722.1 CH916372 EDV99121.1 CM000157 EDW87628.1 KB203854 ESO82760.1 ADMH02000392 ETN66725.1 CH902624 EDV33374.1 CH963920 EDW77828.1 CM000361 EDX03701.1 CH480820 EDW54227.1 AE014134 AY071234 LSMT01000298 PFX20908.1 KK112417 KFM57597.1 EDY70271.2 CM002910 KMY88053.1 IAAA01030192 IAAA01030193 LAA09200.1 GANP01006128 JAB78340.1 CH933807 EDW12976.1 KRG03526.1 NEDP02005575 OWF38030.1 JH816693 EKC29341.1 CP012523 ALC38744.1
KOB67400.1 ODYU01001085 SOQ36827.1 KQ461033 KPJ09973.1 KQ458793 KPJ04971.1 AGBW02012180 OWR45505.1 GAIX01004030 JAA88530.1 KK853292 KDR08843.1 NEVH01000596 PNF43610.1 PYGN01000858 PSN39966.1 GL732545 EFX80914.1 KQ766110 OAD53736.1 DS231895 EDS44358.1 GAMC01000057 JAC06499.1 KQ435922 KOX68504.1 JXUM01010543 KQ560311 KXJ83110.1 CH477400 EAT41746.1 GDAI01002841 JAI14762.1 EZ423897 ADD20173.1 GFDL01015577 JAV19468.1 KZ288300 PBC28887.1 KQ980724 KYN14151.1 KQ434874 KZC09709.1 GEZM01040674 JAV80772.1 KQ976731 KYM76325.1 ADTU01006902 OUUW01000004 SPP79894.1 GL887707 EGI70177.1 KQ978397 KYM94208.1 KQ414582 KOC70914.1 GL442912 EFN62760.1 LRGB01000044 KZS21211.1 KQ982021 KYN30414.1 KQ982892 KYQ49624.1 NNAY01000390 OXU28717.1 GDIP01031145 JAM72570.1 KK107293 QOIP01000005 EZA53178.1 RLU22629.1 GL446361 EFN87940.1 GDHF01007499 JAI44815.1 ATLV01012963 KE524828 KFB37094.1 GAKP01002016 JAC56936.1 GBXI01007410 JAD06882.1 JRES01000438 KNC31322.1 AMQM01003587 KB096222 ESO07225.1 AXCN02002100 APGK01042893 BT127645 KB741009 KB632250 AEE62607.1 ENN75598.1 ERL90592.1 AAAB01006447 EAL42206.2 APCN01001653 GELH01000956 GELH01000955 JAS03317.1 AXCM01000708 KQ971372 EFA10560.2 CH940661 EDW71331.1 CH379060 EDY70270.1 CH479187 EDW39634.1 GEDC01030061 GEDC01022342 JAS07237.1 JAS14956.1 CH954177 EDV57722.1 CH916372 EDV99121.1 CM000157 EDW87628.1 KB203854 ESO82760.1 ADMH02000392 ETN66725.1 CH902624 EDV33374.1 CH963920 EDW77828.1 CM000361 EDX03701.1 CH480820 EDW54227.1 AE014134 AY071234 LSMT01000298 PFX20908.1 KK112417 KFM57597.1 EDY70271.2 CM002910 KMY88053.1 IAAA01030192 IAAA01030193 LAA09200.1 GANP01006128 JAB78340.1 CH933807 EDW12976.1 KRG03526.1 NEDP02005575 OWF38030.1 JH816693 EKC29341.1 CP012523 ALC38744.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000053268
UP000007151
UP000027135
+ More
UP000235965 UP000245037 UP000000305 UP000002320 UP000092445 UP000053105 UP000069940 UP000249989 UP000008820 UP000078200 UP000005203 UP000242457 UP000078492 UP000092443 UP000076502 UP000091820 UP000078540 UP000005205 UP000268350 UP000007755 UP000078542 UP000053825 UP000000311 UP000076858 UP000078541 UP000075809 UP000215335 UP000053097 UP000279307 UP000008237 UP000095300 UP000075880 UP000030765 UP000002358 UP000095301 UP000037069 UP000075920 UP000015101 UP000075886 UP000019118 UP000030742 UP000075903 UP000076407 UP000007062 UP000076408 UP000075882 UP000075840 UP000075902 UP000076420 UP000069272 UP000075883 UP000075900 UP000007266 UP000008792 UP000001819 UP000008744 UP000008711 UP000192221 UP000001070 UP000075884 UP000002282 UP000030746 UP000000673 UP000007801 UP000007798 UP000000304 UP000001292 UP000000803 UP000225706 UP000079169 UP000054359 UP000009192 UP000242188 UP000005408 UP000085678 UP000092553 UP000075885
UP000235965 UP000245037 UP000000305 UP000002320 UP000092445 UP000053105 UP000069940 UP000249989 UP000008820 UP000078200 UP000005203 UP000242457 UP000078492 UP000092443 UP000076502 UP000091820 UP000078540 UP000005205 UP000268350 UP000007755 UP000078542 UP000053825 UP000000311 UP000076858 UP000078541 UP000075809 UP000215335 UP000053097 UP000279307 UP000008237 UP000095300 UP000075880 UP000030765 UP000002358 UP000095301 UP000037069 UP000075920 UP000015101 UP000075886 UP000019118 UP000030742 UP000075903 UP000076407 UP000007062 UP000076408 UP000075882 UP000075840 UP000075902 UP000076420 UP000069272 UP000075883 UP000075900 UP000007266 UP000008792 UP000001819 UP000008744 UP000008711 UP000192221 UP000001070 UP000075884 UP000002282 UP000030746 UP000000673 UP000007801 UP000007798 UP000000304 UP000001292 UP000000803 UP000225706 UP000079169 UP000054359 UP000009192 UP000242188 UP000005408 UP000085678 UP000092553 UP000075885
Interpro
SUPFAM
SSF158573
SSF158573
ProteinModelPortal
H9JAY5
A0A1E1WP58
A0A1C9EGI7
A0A0L7KWA7
A0A2H1V7P2
A0A0N1INJ5
+ More
A0A194QNA9 A0A212EVH9 S4PLH1 A0A067QTG0 A0A2J7RRZ8 A0A2P8Y6T8 E9GHY1 A0A310SFE3 B0WD86 W8C889 A0A1B0A5Y2 A0A0N0BC54 A0A182GEW5 Q175H4 A0A0K8TKA9 D3TR45 A0A1A9V6W8 A0A1Q3EW89 A0A088A6B1 A0A2A3EAY0 A0A195DMV1 A0A1A9X614 A0A154PCQ7 A0A1A9WDL8 A0A1Y1M4S0 A0A195AWB3 A0A158P1W4 A0A3B0JC07 F4W642 A0A151I7X8 A0A0L7RJB2 E2AUX7 A0A162SE06 A0A195ERA8 A0A151WP32 A0A232FEH8 A0A0P6AGA1 A0A026WB65 E2B8N4 A0A1I8QEP2 A0A0K8W1V7 A0A182JKD0 A0A084VGK0 K7J3J2 A0A034WMY9 A0A1I8MQ31 A0A0A1X6Z9 A0A0L0CGI4 A0A182W8N7 T1G2N2 A0A182QKS7 J3JWM7 A0A182VCF8 A0A182X987 Q5TXV1 A0A182Y670 A0A182KZ66 A0A182HR63 A0A182TRK6 A0A2C9JNB1 A0A194ALK1 A0A182F350 A0A182LW41 A0A182RWN4 D6X0V8 B4MDS0 B5DIX7 B4GPW2 A0A1B6CNC0 B3N3R3 A0A1W4WAI5 B4JQE1 A0A182NNZ4 B4NYJ5 V3ZK44 W5JUU6 B3MU45 B4MZP3 B4QA63 B4I382 Q9VQY9 A0A2B4RX52 A0A1S4EMW1 A0A087SXK8 B5DIX8 A0A0J9QVI8 A0A2L2YP27 V5H5P6 B4KES6 A0A210PNE2 K1R5T3 A0A1S3HQ06 A0A0M5J8Z0 A0A182PXD8
A0A194QNA9 A0A212EVH9 S4PLH1 A0A067QTG0 A0A2J7RRZ8 A0A2P8Y6T8 E9GHY1 A0A310SFE3 B0WD86 W8C889 A0A1B0A5Y2 A0A0N0BC54 A0A182GEW5 Q175H4 A0A0K8TKA9 D3TR45 A0A1A9V6W8 A0A1Q3EW89 A0A088A6B1 A0A2A3EAY0 A0A195DMV1 A0A1A9X614 A0A154PCQ7 A0A1A9WDL8 A0A1Y1M4S0 A0A195AWB3 A0A158P1W4 A0A3B0JC07 F4W642 A0A151I7X8 A0A0L7RJB2 E2AUX7 A0A162SE06 A0A195ERA8 A0A151WP32 A0A232FEH8 A0A0P6AGA1 A0A026WB65 E2B8N4 A0A1I8QEP2 A0A0K8W1V7 A0A182JKD0 A0A084VGK0 K7J3J2 A0A034WMY9 A0A1I8MQ31 A0A0A1X6Z9 A0A0L0CGI4 A0A182W8N7 T1G2N2 A0A182QKS7 J3JWM7 A0A182VCF8 A0A182X987 Q5TXV1 A0A182Y670 A0A182KZ66 A0A182HR63 A0A182TRK6 A0A2C9JNB1 A0A194ALK1 A0A182F350 A0A182LW41 A0A182RWN4 D6X0V8 B4MDS0 B5DIX7 B4GPW2 A0A1B6CNC0 B3N3R3 A0A1W4WAI5 B4JQE1 A0A182NNZ4 B4NYJ5 V3ZK44 W5JUU6 B3MU45 B4MZP3 B4QA63 B4I382 Q9VQY9 A0A2B4RX52 A0A1S4EMW1 A0A087SXK8 B5DIX8 A0A0J9QVI8 A0A2L2YP27 V5H5P6 B4KES6 A0A210PNE2 K1R5T3 A0A1S3HQ06 A0A0M5J8Z0 A0A182PXD8
PDB
2Q9Q
E-value=3.28311e-46,
Score=462
Ontologies
GO
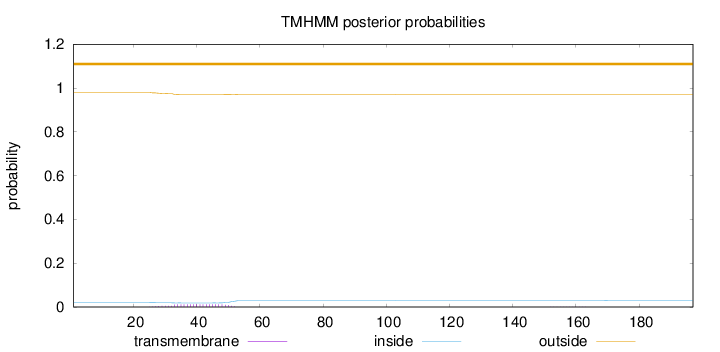
Topology
Subcellular location
Nucleus
Length:
197
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25263
Exp number, first 60 AAs:
0.2501
Total prob of N-in:
0.02032
outside
1 - 197
Population Genetic Test Statistics
Pi
78.148799
Theta
142.008304
Tajima's D
-1.462922
CLR
550.970036
CSRT
0.0675466226688666
Interpretation
Uncertain
