Pre Gene Modal
BGIBMGA006677
Annotation
PREDICTED:_lymphokine-activated_killer_T-cell-originated_protein_kinase_homolog_[Amyelois_transitella]
Full name
Lymphokine-activated killer T-cell-originated protein kinase
Alternative Name
PDZ-binding kinase
T-LAK cell-originated protein kinase
T-LAK cell-originated protein kinase
Location in the cell
Cytoplasmic Reliability : 1.628 Mitochondrial Reliability : 1.001 Nuclear Reliability : 1.172
Sequence
CDS
ATGACCGAATTCCAAACGCCACAAAAGTCTATCAAATCTTTCGACAACGATAAAATAACTATACCACCATCACCGTTTTTAAATAAACTTGGTTATGGTACTGGGGTGTCCGTTCTGCAATTAGTGAGGTCCCCGAGGGCAGGACTTATCCGATCACCGTGGGCTCTCAAAATGTTGAACAAACGAGTTAAACCAAATAAAGTATACACAGATCGACTGAAAACTGAGGCGGAACTGTTACAAAAAATGTCCCACCTTAACATAGTGGGTTTTAGGGCATTTAGTAAAGGCAAGATTCTATACTTGGGCATGGAAGCTTGTGATTTATCCTTGGGGGACTTGATCGAAAAAAGAGTAGATGATGATTGTACGCCCTTTTCACCAAGACAAATGTTGCAGGTGGCTGTAGACATCTCCAGTGCACTTGAATATCTTCATACGAAGATGCAAATTTTACATGGAGACATGAAATCTTACAACATATTAGTCAACGGTGATTTTGTGATATGTAAACTTTGTGACTTTGGAGTGACACTTCCTCTCGACGAAAATGGAATATTTGATAAGGAAAATGCTGGTGGAACTGTTTATTATGGTACAGAAGCATGGAGCGCACCTGAAGTAATTCACGGAGGGCAGATCAGTGACAAAACTGACATCTGGCCATTTGGTCTCACTCTGTGGGAGATGATGTCACTAATGCCACCTCACTCACAAATTGATGAGGACACCTTTGATATGAGCCTGGATGATCTTGACGAGAGTGGTTATGATTTCTCCGATCGATATGGCAGTCGTCCAGACGTCCCCGCCAACATATCGGAAGTAGAATACGCACAGCCGTTAACCCTGTTCTGCAGCTGTACTGAGAGCCGGCCGGCCGACCGCCCCCAGGCCGCGCAGCTCGCGGCGGCCGCCCGGGCCATGGCAGCGGCATTACTTCATAACACATCATGTCAATAG
Protein
MTEFQTPQKSIKSFDNDKITIPPSPFLNKLGYGTGVSVLQLVRSPRAGLIRSPWALKMLNKRVKPNKVYTDRLKTEAELLQKMSHLNIVGFRAFSKGKILYLGMEACDLSLGDLIEKRVDDDCTPFSPRQMLQVAVDISSALEYLHTKMQILHGDMKSYNILVNGDFVICKLCDFGVTLPLDENGIFDKENAGGTVYYGTEAWSAPEVIHGGQISDKTDIWPFGLTLWEMMSLMPPHSQIDEDTFDMSLDDLDESGYDFSDRYGSRPDVPANISEVEYAQPLTLFCSCTESRPADRPQAAQLAAAARAMAAALLHNTSCQ
Summary
Description
Phosphorylates MAP kinase p38. Seems to be active only in mitosis. May also play a role in the activation of lymphoid cells. When phosphorylated, forms a complex with TP53, leading to TP53 destabilization (By similarity).
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with DLG1 and TP53.
Similarity
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase subfamily.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase subfamily.
Keywords
Acetylation
ATP-binding
Complete proteome
Isopeptide bond
Kinase
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Ubl conjugation
Feature
chain Lymphokine-activated killer T-cell-originated protein kinase
Uniprot
H9JAY2
A0A2W1BMY4
A0A194QHU3
A0A2H1WJR3
A0A0N1IFI9
A0A212EUN1
+ More
S4PCQ9 A0A0L7L0W0 A0A1Y1KVP1 A0A088AVR0 A0A2A3ERM6 A0A2J7PPK5 A0A2J7PPN8 A0A0M8ZRD5 A0A154PQA7 N6U5D1 A0A232FIR1 K7IPL6 A0A0C9RA33 A0A0L7RJ96 A0A0A9YN02 A0A146L2V1 W8CAY8 A0A224XPY9 A0A034V6J4 A0A1L8DLP5 A0A067R2H2 B3MQV1 A0A0K8UCY4 E2AD67 Q17F09 A0A0M5J1A0 A0A182G763 A0A1W2W7V1 Q16FK2 B4JKB9 B4MA04 A0A1S4F5F8 A0A0P6ITU8 A0A2G8KW59 A0A0P4VY17 H2Y7F9 A0A336L5Z7 A0A151J1M1 A0A026X1S3 B4L7V2 A0A1B0GIR0 A0A182GX95 B5DKM5 A0A1W4V7M8 A0A1W4V848 B4H9W0 A0A1B6K9T6 A0A1Q3FVN7 A0A1I8Q1S6 A0A1I8N2R6 T1PLZ9 A0A1W4XVF0 A0A0L0BWS0 G3X202 A0A087Y936 A0A1B6CDM0 A0A0J7KVB2 E9G698 A0A3B3Z359 F4WTX2 B3NWX1 A0A2I0LJ26 A0A3B0JZ94 E0V8X5 A0A1B6HCD9 A0A151WN00 Q9JJ78 A0A1A9VRG9 M3XPA9 Q9VX24 R7VP95 A0A2Y9DVV2 A0A2Y9KCT4 A0A195AUR3 A0A158P0Y7 B4Q2J4 A0A2K5E475 B4R6F9 Q5M7R0 A0A1B6GXC0 A0A315W681 A0A3P8USY8 G3SN82 A0A2Y9HXL1 G1PE13 A0A2U3WR42 A0A384D8D2 A0A3Q7Y2R4 M3WKP6 A6QQX0 F7CIG7 L5LSF2 A0A2K6FJT9
S4PCQ9 A0A0L7L0W0 A0A1Y1KVP1 A0A088AVR0 A0A2A3ERM6 A0A2J7PPK5 A0A2J7PPN8 A0A0M8ZRD5 A0A154PQA7 N6U5D1 A0A232FIR1 K7IPL6 A0A0C9RA33 A0A0L7RJ96 A0A0A9YN02 A0A146L2V1 W8CAY8 A0A224XPY9 A0A034V6J4 A0A1L8DLP5 A0A067R2H2 B3MQV1 A0A0K8UCY4 E2AD67 Q17F09 A0A0M5J1A0 A0A182G763 A0A1W2W7V1 Q16FK2 B4JKB9 B4MA04 A0A1S4F5F8 A0A0P6ITU8 A0A2G8KW59 A0A0P4VY17 H2Y7F9 A0A336L5Z7 A0A151J1M1 A0A026X1S3 B4L7V2 A0A1B0GIR0 A0A182GX95 B5DKM5 A0A1W4V7M8 A0A1W4V848 B4H9W0 A0A1B6K9T6 A0A1Q3FVN7 A0A1I8Q1S6 A0A1I8N2R6 T1PLZ9 A0A1W4XVF0 A0A0L0BWS0 G3X202 A0A087Y936 A0A1B6CDM0 A0A0J7KVB2 E9G698 A0A3B3Z359 F4WTX2 B3NWX1 A0A2I0LJ26 A0A3B0JZ94 E0V8X5 A0A1B6HCD9 A0A151WN00 Q9JJ78 A0A1A9VRG9 M3XPA9 Q9VX24 R7VP95 A0A2Y9DVV2 A0A2Y9KCT4 A0A195AUR3 A0A158P0Y7 B4Q2J4 A0A2K5E475 B4R6F9 Q5M7R0 A0A1B6GXC0 A0A315W681 A0A3P8USY8 G3SN82 A0A2Y9HXL1 G1PE13 A0A2U3WR42 A0A384D8D2 A0A3Q7Y2R4 M3WKP6 A6QQX0 F7CIG7 L5LSF2 A0A2K6FJT9
EC Number
2.7.12.2
Pubmed
19121390
28756777
26354079
22118469
23622113
26227816
+ More
28004739 23537049 28648823 20075255 25401762 26823975 24495485 25348373 24845553 17994087 20798317 17510324 26483478 12481130 15114417 26999592 29023486 24508170 30249741 15632085 25315136 26108605 21709235 21292972 21719571 23371554 20566863 10781613 16141072 15489334 21183079 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 17550304 29703783 24487278 21993624 24813606 17975172 19393038 20431018
28004739 23537049 28648823 20075255 25401762 26823975 24495485 25348373 24845553 17994087 20798317 17510324 26483478 12481130 15114417 26999592 29023486 24508170 30249741 15632085 25315136 26108605 21709235 21292972 21719571 23371554 20566863 10781613 16141072 15489334 21183079 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 17550304 29703783 24487278 21993624 24813606 17975172 19393038 20431018
EMBL
BABH01008264
KZ150036
PZC74637.1
KQ458793
KPJ04964.1
ODYU01009124
+ More
SOQ53325.1 KQ461033 KPJ09963.1 AGBW02012335 OWR45193.1 GAIX01004041 JAA88519.1 JTDY01003694 KOB69133.1 GEZM01074210 JAV64638.1 KZ288198 PBC33691.1 NEVH01022660 PNF18265.1 PNF18266.1 KQ435922 KOX68426.1 KQ434998 KZC13300.1 APGK01041766 KB740997 KB632100 ENN75856.1 ERL88837.1 NNAY01000187 OXU30187.1 GBYB01000849 GBYB01000850 GBYB01005100 JAG70616.1 JAG70617.1 JAG74867.1 KQ414582 KOC70858.1 GBHO01009172 GBRD01012327 GBRD01011978 JAG34432.1 JAG53497.1 GDHC01015775 JAQ02854.1 GAMC01000939 JAC05617.1 GFTR01004528 JAW11898.1 GAKP01021774 JAC37178.1 GFDF01006696 JAV07388.1 KK853060 KDR11913.1 CH902622 EDV34156.1 GDHF01027878 GDHF01004460 JAI24436.1 JAI47854.1 GL438684 EFN68615.1 CH477277 EAT45123.1 CP012528 ALC49394.1 JXUM01007303 KQ560237 KXJ83674.1 EAAA01002898 CH478422 EAT33016.1 CH916370 EDW00022.1 CH940655 EDW66063.1 GDUN01000759 JAN95160.1 MRZV01000337 PIK52247.1 GDRN01088409 JAI60857.1 UFQS01001726 UFQS01002209 UFQT01001726 UFQT01002209 SSX12019.1 SSX13499.1 SSX31537.1 KQ980514 KYN15741.1 KK107034 QOIP01000008 EZA62023.1 RLU19608.1 CH933814 EDW05527.1 AJWK01015302 JXUM01094873 KQ564159 KXJ72657.1 CH379063 EDY72075.1 CH479232 EDW36617.1 GEBQ01031792 JAT08185.1 GFDL01003398 JAV31647.1 KA648943 AFP63572.1 JRES01001304 KNC23699.1 AEFK01024940 AEFK01024941 AYCK01000193 GEDC01025736 JAS11562.1 LBMM01002886 KMQ94241.1 GL732533 EFX84896.1 GL888345 EGI62329.1 CH954180 EDV46518.1 AKCR02000318 PKK17442.1 OUUW01000003 SPP78696.1 DS234986 EEB09831.1 GECU01035405 JAS72301.1 KQ982934 KYQ49190.1 AB041882 AK003838 AK076121 BC006754 BC020099 AEYP01085699 AE014298 AY071211 AAF48758.1 AAL48833.1 KB375946 EMC79729.1 KQ976738 KYM75760.1 ADTU01005903 CM000162 EDX02635.1 CM000366 EDX18209.1 BC088502 CR761713 AAH88502.1 CAJ83951.1 GECZ01002706 JAS67063.1 NHOQ01001000 PWA27342.1 AAPE02002005 AANG04001605 BC150025 AAI50026.1 AAMC01115299 KB108036 ELK29384.1
SOQ53325.1 KQ461033 KPJ09963.1 AGBW02012335 OWR45193.1 GAIX01004041 JAA88519.1 JTDY01003694 KOB69133.1 GEZM01074210 JAV64638.1 KZ288198 PBC33691.1 NEVH01022660 PNF18265.1 PNF18266.1 KQ435922 KOX68426.1 KQ434998 KZC13300.1 APGK01041766 KB740997 KB632100 ENN75856.1 ERL88837.1 NNAY01000187 OXU30187.1 GBYB01000849 GBYB01000850 GBYB01005100 JAG70616.1 JAG70617.1 JAG74867.1 KQ414582 KOC70858.1 GBHO01009172 GBRD01012327 GBRD01011978 JAG34432.1 JAG53497.1 GDHC01015775 JAQ02854.1 GAMC01000939 JAC05617.1 GFTR01004528 JAW11898.1 GAKP01021774 JAC37178.1 GFDF01006696 JAV07388.1 KK853060 KDR11913.1 CH902622 EDV34156.1 GDHF01027878 GDHF01004460 JAI24436.1 JAI47854.1 GL438684 EFN68615.1 CH477277 EAT45123.1 CP012528 ALC49394.1 JXUM01007303 KQ560237 KXJ83674.1 EAAA01002898 CH478422 EAT33016.1 CH916370 EDW00022.1 CH940655 EDW66063.1 GDUN01000759 JAN95160.1 MRZV01000337 PIK52247.1 GDRN01088409 JAI60857.1 UFQS01001726 UFQS01002209 UFQT01001726 UFQT01002209 SSX12019.1 SSX13499.1 SSX31537.1 KQ980514 KYN15741.1 KK107034 QOIP01000008 EZA62023.1 RLU19608.1 CH933814 EDW05527.1 AJWK01015302 JXUM01094873 KQ564159 KXJ72657.1 CH379063 EDY72075.1 CH479232 EDW36617.1 GEBQ01031792 JAT08185.1 GFDL01003398 JAV31647.1 KA648943 AFP63572.1 JRES01001304 KNC23699.1 AEFK01024940 AEFK01024941 AYCK01000193 GEDC01025736 JAS11562.1 LBMM01002886 KMQ94241.1 GL732533 EFX84896.1 GL888345 EGI62329.1 CH954180 EDV46518.1 AKCR02000318 PKK17442.1 OUUW01000003 SPP78696.1 DS234986 EEB09831.1 GECU01035405 JAS72301.1 KQ982934 KYQ49190.1 AB041882 AK003838 AK076121 BC006754 BC020099 AEYP01085699 AE014298 AY071211 AAF48758.1 AAL48833.1 KB375946 EMC79729.1 KQ976738 KYM75760.1 ADTU01005903 CM000162 EDX02635.1 CM000366 EDX18209.1 BC088502 CR761713 AAH88502.1 CAJ83951.1 GECZ01002706 JAS67063.1 NHOQ01001000 PWA27342.1 AAPE02002005 AANG04001605 BC150025 AAI50026.1 AAMC01115299 KB108036 ELK29384.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000005203
+ More
UP000242457 UP000235965 UP000053105 UP000076502 UP000019118 UP000030742 UP000215335 UP000002358 UP000053825 UP000027135 UP000007801 UP000000311 UP000008820 UP000092553 UP000069940 UP000249989 UP000008144 UP000001070 UP000008792 UP000230750 UP000007875 UP000078492 UP000053097 UP000279307 UP000009192 UP000092461 UP000001819 UP000192221 UP000008744 UP000095300 UP000095301 UP000192223 UP000037069 UP000007648 UP000028760 UP000036403 UP000000305 UP000261480 UP000007755 UP000008711 UP000053872 UP000268350 UP000009046 UP000075809 UP000000589 UP000078200 UP000000715 UP000000803 UP000248480 UP000248482 UP000078540 UP000005205 UP000002282 UP000233020 UP000000304 UP000265120 UP000007646 UP000248481 UP000001074 UP000245340 UP000261680 UP000291021 UP000286642 UP000011712 UP000009136 UP000008143 UP000233160
UP000242457 UP000235965 UP000053105 UP000076502 UP000019118 UP000030742 UP000215335 UP000002358 UP000053825 UP000027135 UP000007801 UP000000311 UP000008820 UP000092553 UP000069940 UP000249989 UP000008144 UP000001070 UP000008792 UP000230750 UP000007875 UP000078492 UP000053097 UP000279307 UP000009192 UP000092461 UP000001819 UP000192221 UP000008744 UP000095300 UP000095301 UP000192223 UP000037069 UP000007648 UP000028760 UP000036403 UP000000305 UP000261480 UP000007755 UP000008711 UP000053872 UP000268350 UP000009046 UP000075809 UP000000589 UP000078200 UP000000715 UP000000803 UP000248480 UP000248482 UP000078540 UP000005205 UP000002282 UP000233020 UP000000304 UP000265120 UP000007646 UP000248481 UP000001074 UP000245340 UP000261680 UP000291021 UP000286642 UP000011712 UP000009136 UP000008143 UP000233160
Interpro
CDD
ProteinModelPortal
H9JAY2
A0A2W1BMY4
A0A194QHU3
A0A2H1WJR3
A0A0N1IFI9
A0A212EUN1
+ More
S4PCQ9 A0A0L7L0W0 A0A1Y1KVP1 A0A088AVR0 A0A2A3ERM6 A0A2J7PPK5 A0A2J7PPN8 A0A0M8ZRD5 A0A154PQA7 N6U5D1 A0A232FIR1 K7IPL6 A0A0C9RA33 A0A0L7RJ96 A0A0A9YN02 A0A146L2V1 W8CAY8 A0A224XPY9 A0A034V6J4 A0A1L8DLP5 A0A067R2H2 B3MQV1 A0A0K8UCY4 E2AD67 Q17F09 A0A0M5J1A0 A0A182G763 A0A1W2W7V1 Q16FK2 B4JKB9 B4MA04 A0A1S4F5F8 A0A0P6ITU8 A0A2G8KW59 A0A0P4VY17 H2Y7F9 A0A336L5Z7 A0A151J1M1 A0A026X1S3 B4L7V2 A0A1B0GIR0 A0A182GX95 B5DKM5 A0A1W4V7M8 A0A1W4V848 B4H9W0 A0A1B6K9T6 A0A1Q3FVN7 A0A1I8Q1S6 A0A1I8N2R6 T1PLZ9 A0A1W4XVF0 A0A0L0BWS0 G3X202 A0A087Y936 A0A1B6CDM0 A0A0J7KVB2 E9G698 A0A3B3Z359 F4WTX2 B3NWX1 A0A2I0LJ26 A0A3B0JZ94 E0V8X5 A0A1B6HCD9 A0A151WN00 Q9JJ78 A0A1A9VRG9 M3XPA9 Q9VX24 R7VP95 A0A2Y9DVV2 A0A2Y9KCT4 A0A195AUR3 A0A158P0Y7 B4Q2J4 A0A2K5E475 B4R6F9 Q5M7R0 A0A1B6GXC0 A0A315W681 A0A3P8USY8 G3SN82 A0A2Y9HXL1 G1PE13 A0A2U3WR42 A0A384D8D2 A0A3Q7Y2R4 M3WKP6 A6QQX0 F7CIG7 L5LSF2 A0A2K6FJT9
S4PCQ9 A0A0L7L0W0 A0A1Y1KVP1 A0A088AVR0 A0A2A3ERM6 A0A2J7PPK5 A0A2J7PPN8 A0A0M8ZRD5 A0A154PQA7 N6U5D1 A0A232FIR1 K7IPL6 A0A0C9RA33 A0A0L7RJ96 A0A0A9YN02 A0A146L2V1 W8CAY8 A0A224XPY9 A0A034V6J4 A0A1L8DLP5 A0A067R2H2 B3MQV1 A0A0K8UCY4 E2AD67 Q17F09 A0A0M5J1A0 A0A182G763 A0A1W2W7V1 Q16FK2 B4JKB9 B4MA04 A0A1S4F5F8 A0A0P6ITU8 A0A2G8KW59 A0A0P4VY17 H2Y7F9 A0A336L5Z7 A0A151J1M1 A0A026X1S3 B4L7V2 A0A1B0GIR0 A0A182GX95 B5DKM5 A0A1W4V7M8 A0A1W4V848 B4H9W0 A0A1B6K9T6 A0A1Q3FVN7 A0A1I8Q1S6 A0A1I8N2R6 T1PLZ9 A0A1W4XVF0 A0A0L0BWS0 G3X202 A0A087Y936 A0A1B6CDM0 A0A0J7KVB2 E9G698 A0A3B3Z359 F4WTX2 B3NWX1 A0A2I0LJ26 A0A3B0JZ94 E0V8X5 A0A1B6HCD9 A0A151WN00 Q9JJ78 A0A1A9VRG9 M3XPA9 Q9VX24 R7VP95 A0A2Y9DVV2 A0A2Y9KCT4 A0A195AUR3 A0A158P0Y7 B4Q2J4 A0A2K5E475 B4R6F9 Q5M7R0 A0A1B6GXC0 A0A315W681 A0A3P8USY8 G3SN82 A0A2Y9HXL1 G1PE13 A0A2U3WR42 A0A384D8D2 A0A3Q7Y2R4 M3WKP6 A6QQX0 F7CIG7 L5LSF2 A0A2K6FJT9
PDB
5J0A
E-value=5.94275e-53,
Score=524
Ontologies
GO
PANTHER
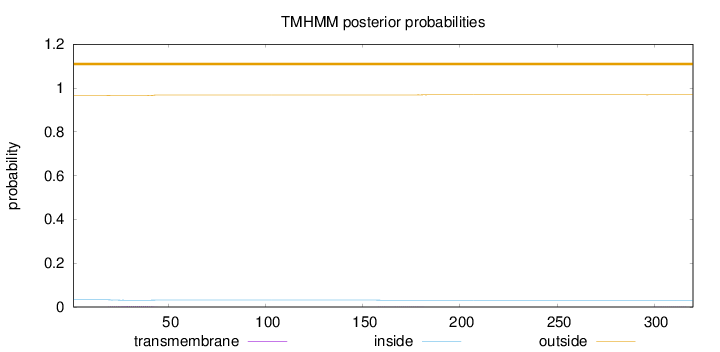
Topology
Length:
320
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0711300000000001
Exp number, first 60 AAs:
0.05478
Total prob of N-in:
0.03324
outside
1 - 320
Population Genetic Test Statistics
Pi
217.06634
Theta
177.685449
Tajima's D
0.697281
CLR
0.00023
CSRT
0.569771511424429
Interpretation
Uncertain
