Gene
KWMTBOMO05399
Pre Gene Modal
BGIBMGA006676
Annotation
PREDICTED:_nuclear_receptor-binding_protein_homolog_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.643 Nuclear Reliability : 1.956
Sequence
CDS
ATGGTATCTTTAGCCGAAGTTGAACAACGCGATGTGCCCGGTATTGACTGCGCCAACCTTGCGATGGACACCGAAGAGGGCGTGGAAGTCGTGTGGAATGAAGTCCAGTTCTCCGAGCGGAAGAACTTCAAAGCTCAGGAAGATAAGATACAGACGGTGTTCGACAACCTGACCAGGCTTGAACACCCTAACATCGTCAAGTTCCATCGATACTGGACTGACACGCATAACGACAAACCCAGGGTAATTTTCATTACTGAGTACATGTCGTGCGGCTCATTGAAGCAGTTTCTGAAGAGAACGAAGCGCAACGTGAAGAGGCTGCCCCTCCAGGCGTGGAAGCGATGGTGCACACAGATACTATCTGCATTAAGTTATCTCCACGGCTGTGTTCCGCCGATCGTCCACGGAAACCTGACCTGCGACACCATTTTCATCCAGCATAACGGCCTGGTGAAGATCGGCTCTGTAGCGCCCGACGCAATTCACCACCACGTCAAGACCTGCCGCGAAAATATGAAGAACATGCACTTTATCGCCCCAGAATATGGATCATCTCAAATGGTGACGCCAGCAATGGATATCTACTCGTTCGGAATGTGCGCTCTGGAAACCGCAGCCCTCGAAATCCAAGGAAATGGAGACTCTGGCAGCCACGTCACGGAGGAACACATCATACGGACGGTCGAATCGTTGGAGGATCCCAGACAGAAGGACTTTATATACAGGTGTATAAACCAGCATCCCGAGAAGAGGCCGACGGCGAGAGAGCTGCTCTTCCATCCACTGCTGTTTGAAGTCCATTCCTTAAAACTGCTGGCGGCCCACACGCTGGTTAATACAACCGCTAATATCTCCGAAACTATAACTGACGAAGTGACCGGCGGCAGCAGCGGCGAGGCGCTGGCGTGGTGCGTGCGCGGCGGGGAGCGCGTCGAGATCGCGGCCAAGGACCTCCAGCCGCACGCTGAAAAACTAGAGAAATTTGTCGAAGACGTCAAGTACGGCATCTACCCGCTGACGGCGTACGGGTCGGGGGCGGGGGGCCGCGCCTCGTCCCCCGAGCGCGCCCCCGCCCCCGCCGCCTCGCCCCCCCCGCGCGACACCGAGTCCCGCCGCGTCGTGCACATGATGTGCAACGTGGCGCCGCCGCCGCGACACAGCGACGCGCCCGACCACGACCTGCTGATGACCATCATCCTGCGCATGGACGACAAGATGAACCGGCAGCTGACGTGCGCCGTGTCGCGCCGGGACTCCGCGTCGCACCTCGCGCGCGAGCTGGTGCAGCTCGGCTTCATCCACGAGGCGGACCGCGACAAGATCTGCCGGATCATGGAGGAGTCCCTCCGCGCCAGTTTCCCCCCGGCGGCGGAGCCCCCCGTGGCGGCCCCCCCGGCGGCGGCGGCCCGCGCGCCTCAGCACGTGGCCAGCTAG
Protein
MVSLAEVEQRDVPGIDCANLAMDTEEGVEVVWNEVQFSERKNFKAQEDKIQTVFDNLTRLEHPNIVKFHRYWTDTHNDKPRVIFITEYMSCGSLKQFLKRTKRNVKRLPLQAWKRWCTQILSALSYLHGCVPPIVHGNLTCDTIFIQHNGLVKIGSVAPDAIHHHVKTCRENMKNMHFIAPEYGSSQMVTPAMDIYSFGMCALETAALEIQGNGDSGSHVTEEHIIRTVESLEDPRQKDFIYRCINQHPEKRPTARELLFHPLLFEVHSLKLLAAHTLVNTTANISETITDEVTGGSSGEALAWCVRGGERVEIAAKDLQPHAEKLEKFVEDVKYGIYPLTAYGSGAGGRASSPERAPAPAASPPPRDTESRRVVHMMCNVAPPPRHSDAPDHDLLMTIILRMDDKMNRQLTCAVSRRDSASHLARELVQLGFIHEADRDKICRIMEESLRASFPPAAEPPVAAPPAAAARAPQHVAS
Summary
Uniprot
A0A2H1W7S7
H9JAY1
A0A0N1INJ3
A0A2W1BLU6
A0A3S2NMN9
A0A212FKZ1
+ More
A0A0K8TKC3 A0A195CF60 A0A195D9T1 F4W503 A0A195BNZ3 U5ES75 A0A154PT47 A0A195F4A6 E2BQD9 E1ZWL4 E0V8X1 E9J308 A0A151XEB3 A0A2A3EN58 A0A2A3ELQ3 A0A088A122 A0A1L8DLC5 A0A0C9R8P4 A0A0C9QHM4 A0A0C9R5E9 A0A232EN53 K7IX41 A0A0L7QUA7 A0A1B6L6Y6 A0A067QQ77 A0A1B6EKZ8 A0A3L8DN88 A0A1J1I227 A0A2M4A3W5 A0A2M4CTL3 A0A2M4CZ05 A0A182F1L1 A0A1B6I0C3 A0A2M3Z328 A0A1B0CQK4 A0A069DZA0 W5JSU5 A0A0V0GDZ3 A0A023F5B9 A0A0A9WT01 T1DJH1 A0A146L9D2 A0A1Q3FX82 A0A1Q3FWU9 A0A0J7KSP5 A0A2J7PPL7 A0A1B6CA41 A0A182G4R9 A0A1Y1N5W8 Q17JY5 A0A139WCG3 A0A182L1V2 A0A1S4GXM4 A0A1S4GWY5 A0A1S4J236 A0A084WD80 A0A0N0U6W4 A0A182KA85 A0A182W7E6 Q7Q3R5 A0A182NTM5 A0A1S4EZX7 A0A182HV10 A0A2S2NY27 X1XCS2 A0A1D2N5X5 E9H8B9 A0A0N8CEL9 A0A0P5LVB8 A0A224YWX5 A0A131YI29 A0A182XZX4 A0A293MXU7 A0A087TCA0 A0A0P5A281 A0A0P5E814 A0A0P5HRR2 A0A0P5SS50 A0A162TAP3 A0A0P5BVI6 A0A0P5K519 A0A0P5HNG8 A0A182LZS9 A0A1Z5KW83 A0A182QD76 A0A0P5DRK5 A0A0P5YKY0 A0A0P6ED54 A0A0P5Y1W3 A0A0P5VD78 A0A0P5VV19 A0A0P5HXL1 A0A1E1XQI5 A0A023GK87
A0A0K8TKC3 A0A195CF60 A0A195D9T1 F4W503 A0A195BNZ3 U5ES75 A0A154PT47 A0A195F4A6 E2BQD9 E1ZWL4 E0V8X1 E9J308 A0A151XEB3 A0A2A3EN58 A0A2A3ELQ3 A0A088A122 A0A1L8DLC5 A0A0C9R8P4 A0A0C9QHM4 A0A0C9R5E9 A0A232EN53 K7IX41 A0A0L7QUA7 A0A1B6L6Y6 A0A067QQ77 A0A1B6EKZ8 A0A3L8DN88 A0A1J1I227 A0A2M4A3W5 A0A2M4CTL3 A0A2M4CZ05 A0A182F1L1 A0A1B6I0C3 A0A2M3Z328 A0A1B0CQK4 A0A069DZA0 W5JSU5 A0A0V0GDZ3 A0A023F5B9 A0A0A9WT01 T1DJH1 A0A146L9D2 A0A1Q3FX82 A0A1Q3FWU9 A0A0J7KSP5 A0A2J7PPL7 A0A1B6CA41 A0A182G4R9 A0A1Y1N5W8 Q17JY5 A0A139WCG3 A0A182L1V2 A0A1S4GXM4 A0A1S4GWY5 A0A1S4J236 A0A084WD80 A0A0N0U6W4 A0A182KA85 A0A182W7E6 Q7Q3R5 A0A182NTM5 A0A1S4EZX7 A0A182HV10 A0A2S2NY27 X1XCS2 A0A1D2N5X5 E9H8B9 A0A0N8CEL9 A0A0P5LVB8 A0A224YWX5 A0A131YI29 A0A182XZX4 A0A293MXU7 A0A087TCA0 A0A0P5A281 A0A0P5E814 A0A0P5HRR2 A0A0P5SS50 A0A162TAP3 A0A0P5BVI6 A0A0P5K519 A0A0P5HNG8 A0A182LZS9 A0A1Z5KW83 A0A182QD76 A0A0P5DRK5 A0A0P5YKY0 A0A0P6ED54 A0A0P5Y1W3 A0A0P5VD78 A0A0P5VV19 A0A0P5HXL1 A0A1E1XQI5 A0A023GK87
Pubmed
19121390
26354079
28756777
22118469
26369729
21719571
+ More
20798317 20566863 21282665 28648823 20075255 24845553 30249741 26334808 20920257 23761445 25474469 25401762 24330624 26823975 26483478 28004739 17510324 18362917 19820115 20966253 24438588 12364791 27289101 21292972 28797301 26830274 25244985 28528879 29209593
20798317 20566863 21282665 28648823 20075255 24845553 30249741 26334808 20920257 23761445 25474469 25401762 24330624 26823975 26483478 28004739 17510324 18362917 19820115 20966253 24438588 12364791 27289101 21292972 28797301 26830274 25244985 28528879 29209593
EMBL
ODYU01006811
SOQ49006.1
BABH01008260
BABH01008261
KQ461033
KPJ09960.1
+ More
KZ150036 PZC74635.1 RSAL01000032 RVE51563.1 AGBW02007931 OWR54413.1 GDAI01002894 JAI14709.1 KQ977827 KYM99432.1 KQ981082 KYN09638.1 GL887596 EGI70699.1 KQ976424 KYM88237.1 GANO01003375 JAB56496.1 KQ435078 KZC14504.1 KQ981820 KYN35216.1 GL449738 EFN82103.1 GL434853 EFN74451.1 DS234986 EEB09827.1 GL768055 EFZ12814.1 KQ982254 KYQ58712.1 KZ288215 PBC32636.1 PBC32637.1 GFDF01006815 JAV07269.1 GBYB01003161 JAG72928.1 GBYB01003029 GBYB01003162 JAG72796.1 JAG72929.1 GBYB01003165 JAG72932.1 NNAY01003236 OXU19757.1 AAZX01000620 KQ414735 KOC62242.1 GEBQ01020489 JAT19488.1 KK853060 KDR11910.1 GECZ01031155 JAS38614.1 QOIP01000006 RLU21867.1 CVRI01000038 CRK93818.1 GGFK01002128 MBW35449.1 GGFL01004518 MBW68696.1 GGFL01006386 MBW70564.1 GECU01027340 GECU01021368 GECU01014762 JAS80366.1 JAS86338.1 JAS92944.1 GGFM01002154 MBW22905.1 AJWK01023708 AJWK01023709 AJWK01023710 GBGD01001005 JAC87884.1 ADMH02000259 ETN67216.1 GECL01000535 JAP05589.1 GBBI01002403 JAC16309.1 GBHO01035624 GBRD01016238 JAG07980.1 JAG49588.1 GALA01000266 JAA94586.1 GDHC01013685 JAQ04944.1 GFDL01003012 JAV32033.1 GFDL01003010 JAV32035.1 LBMM01003522 KMQ93422.1 NEVH01022660 PNF18246.1 GEDC01026977 JAS10321.1 JXUM01042525 JXUM01042526 KQ561328 KXJ78963.1 GEZM01012212 JAV93099.1 CH477229 EAT47015.1 KQ971371 KYB25545.1 ATLV01022955 KE525338 KFB48174.1 KQ435716 KOX79024.1 AAAB01008964 EAA12395.4 APCN01002488 GGMR01009466 MBY22085.1 ABLF02031121 LJIJ01000194 ODN00657.1 GL732603 EFX72053.1 GDIP01139295 JAL64419.1 GDIQ01189164 JAK62561.1 GFPF01010971 MAA22117.1 GEDV01009613 JAP78944.1 GFWV01020221 MAA44949.1 KK114562 KFM62739.1 GDIP01205332 JAJ18070.1 GDIP01151388 JAJ72014.1 GDIQ01224193 JAK27532.1 GDIQ01204754 GDIP01139296 JAK46971.1 JAL64418.1 LRGB01000005 KZS22044.1 GDIP01179887 JAJ43515.1 GDIQ01189166 JAK62559.1 GDIQ01225448 JAK26277.1 AXCM01008540 GFJQ02007604 JAV99365.1 AXCN02000795 GDIP01153844 JAJ69558.1 GDIP01056344 JAM47371.1 GDIQ01064457 JAN30280.1 GDIP01064106 JAM39609.1 GDIP01116781 JAL86933.1 GDIP01095446 JAM08269.1 GDIQ01221389 JAK30336.1 GFAA01001844 JAU01591.1 GBBM01001074 JAC34344.1
KZ150036 PZC74635.1 RSAL01000032 RVE51563.1 AGBW02007931 OWR54413.1 GDAI01002894 JAI14709.1 KQ977827 KYM99432.1 KQ981082 KYN09638.1 GL887596 EGI70699.1 KQ976424 KYM88237.1 GANO01003375 JAB56496.1 KQ435078 KZC14504.1 KQ981820 KYN35216.1 GL449738 EFN82103.1 GL434853 EFN74451.1 DS234986 EEB09827.1 GL768055 EFZ12814.1 KQ982254 KYQ58712.1 KZ288215 PBC32636.1 PBC32637.1 GFDF01006815 JAV07269.1 GBYB01003161 JAG72928.1 GBYB01003029 GBYB01003162 JAG72796.1 JAG72929.1 GBYB01003165 JAG72932.1 NNAY01003236 OXU19757.1 AAZX01000620 KQ414735 KOC62242.1 GEBQ01020489 JAT19488.1 KK853060 KDR11910.1 GECZ01031155 JAS38614.1 QOIP01000006 RLU21867.1 CVRI01000038 CRK93818.1 GGFK01002128 MBW35449.1 GGFL01004518 MBW68696.1 GGFL01006386 MBW70564.1 GECU01027340 GECU01021368 GECU01014762 JAS80366.1 JAS86338.1 JAS92944.1 GGFM01002154 MBW22905.1 AJWK01023708 AJWK01023709 AJWK01023710 GBGD01001005 JAC87884.1 ADMH02000259 ETN67216.1 GECL01000535 JAP05589.1 GBBI01002403 JAC16309.1 GBHO01035624 GBRD01016238 JAG07980.1 JAG49588.1 GALA01000266 JAA94586.1 GDHC01013685 JAQ04944.1 GFDL01003012 JAV32033.1 GFDL01003010 JAV32035.1 LBMM01003522 KMQ93422.1 NEVH01022660 PNF18246.1 GEDC01026977 JAS10321.1 JXUM01042525 JXUM01042526 KQ561328 KXJ78963.1 GEZM01012212 JAV93099.1 CH477229 EAT47015.1 KQ971371 KYB25545.1 ATLV01022955 KE525338 KFB48174.1 KQ435716 KOX79024.1 AAAB01008964 EAA12395.4 APCN01002488 GGMR01009466 MBY22085.1 ABLF02031121 LJIJ01000194 ODN00657.1 GL732603 EFX72053.1 GDIP01139295 JAL64419.1 GDIQ01189164 JAK62561.1 GFPF01010971 MAA22117.1 GEDV01009613 JAP78944.1 GFWV01020221 MAA44949.1 KK114562 KFM62739.1 GDIP01205332 JAJ18070.1 GDIP01151388 JAJ72014.1 GDIQ01224193 JAK27532.1 GDIQ01204754 GDIP01139296 JAK46971.1 JAL64418.1 LRGB01000005 KZS22044.1 GDIP01179887 JAJ43515.1 GDIQ01189166 JAK62559.1 GDIQ01225448 JAK26277.1 AXCM01008540 GFJQ02007604 JAV99365.1 AXCN02000795 GDIP01153844 JAJ69558.1 GDIP01056344 JAM47371.1 GDIQ01064457 JAN30280.1 GDIP01064106 JAM39609.1 GDIP01116781 JAL86933.1 GDIP01095446 JAM08269.1 GDIQ01221389 JAK30336.1 GFAA01001844 JAU01591.1 GBBM01001074 JAC34344.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000007151
UP000078542
UP000078492
+ More
UP000007755 UP000078540 UP000076502 UP000078541 UP000008237 UP000000311 UP000009046 UP000075809 UP000242457 UP000005203 UP000215335 UP000002358 UP000053825 UP000027135 UP000279307 UP000183832 UP000069272 UP000092461 UP000000673 UP000036403 UP000235965 UP000069940 UP000249989 UP000008820 UP000007266 UP000075882 UP000030765 UP000053105 UP000075881 UP000075920 UP000007062 UP000075884 UP000075840 UP000007819 UP000094527 UP000000305 UP000076408 UP000054359 UP000076858 UP000075883 UP000075886
UP000007755 UP000078540 UP000076502 UP000078541 UP000008237 UP000000311 UP000009046 UP000075809 UP000242457 UP000005203 UP000215335 UP000002358 UP000053825 UP000027135 UP000279307 UP000183832 UP000069272 UP000092461 UP000000673 UP000036403 UP000235965 UP000069940 UP000249989 UP000008820 UP000007266 UP000075882 UP000030765 UP000053105 UP000075881 UP000075920 UP000007062 UP000075884 UP000075840 UP000007819 UP000094527 UP000000305 UP000076408 UP000054359 UP000076858 UP000075883 UP000075886
PRIDE
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2H1W7S7
H9JAY1
A0A0N1INJ3
A0A2W1BLU6
A0A3S2NMN9
A0A212FKZ1
+ More
A0A0K8TKC3 A0A195CF60 A0A195D9T1 F4W503 A0A195BNZ3 U5ES75 A0A154PT47 A0A195F4A6 E2BQD9 E1ZWL4 E0V8X1 E9J308 A0A151XEB3 A0A2A3EN58 A0A2A3ELQ3 A0A088A122 A0A1L8DLC5 A0A0C9R8P4 A0A0C9QHM4 A0A0C9R5E9 A0A232EN53 K7IX41 A0A0L7QUA7 A0A1B6L6Y6 A0A067QQ77 A0A1B6EKZ8 A0A3L8DN88 A0A1J1I227 A0A2M4A3W5 A0A2M4CTL3 A0A2M4CZ05 A0A182F1L1 A0A1B6I0C3 A0A2M3Z328 A0A1B0CQK4 A0A069DZA0 W5JSU5 A0A0V0GDZ3 A0A023F5B9 A0A0A9WT01 T1DJH1 A0A146L9D2 A0A1Q3FX82 A0A1Q3FWU9 A0A0J7KSP5 A0A2J7PPL7 A0A1B6CA41 A0A182G4R9 A0A1Y1N5W8 Q17JY5 A0A139WCG3 A0A182L1V2 A0A1S4GXM4 A0A1S4GWY5 A0A1S4J236 A0A084WD80 A0A0N0U6W4 A0A182KA85 A0A182W7E6 Q7Q3R5 A0A182NTM5 A0A1S4EZX7 A0A182HV10 A0A2S2NY27 X1XCS2 A0A1D2N5X5 E9H8B9 A0A0N8CEL9 A0A0P5LVB8 A0A224YWX5 A0A131YI29 A0A182XZX4 A0A293MXU7 A0A087TCA0 A0A0P5A281 A0A0P5E814 A0A0P5HRR2 A0A0P5SS50 A0A162TAP3 A0A0P5BVI6 A0A0P5K519 A0A0P5HNG8 A0A182LZS9 A0A1Z5KW83 A0A182QD76 A0A0P5DRK5 A0A0P5YKY0 A0A0P6ED54 A0A0P5Y1W3 A0A0P5VD78 A0A0P5VV19 A0A0P5HXL1 A0A1E1XQI5 A0A023GK87
A0A0K8TKC3 A0A195CF60 A0A195D9T1 F4W503 A0A195BNZ3 U5ES75 A0A154PT47 A0A195F4A6 E2BQD9 E1ZWL4 E0V8X1 E9J308 A0A151XEB3 A0A2A3EN58 A0A2A3ELQ3 A0A088A122 A0A1L8DLC5 A0A0C9R8P4 A0A0C9QHM4 A0A0C9R5E9 A0A232EN53 K7IX41 A0A0L7QUA7 A0A1B6L6Y6 A0A067QQ77 A0A1B6EKZ8 A0A3L8DN88 A0A1J1I227 A0A2M4A3W5 A0A2M4CTL3 A0A2M4CZ05 A0A182F1L1 A0A1B6I0C3 A0A2M3Z328 A0A1B0CQK4 A0A069DZA0 W5JSU5 A0A0V0GDZ3 A0A023F5B9 A0A0A9WT01 T1DJH1 A0A146L9D2 A0A1Q3FX82 A0A1Q3FWU9 A0A0J7KSP5 A0A2J7PPL7 A0A1B6CA41 A0A182G4R9 A0A1Y1N5W8 Q17JY5 A0A139WCG3 A0A182L1V2 A0A1S4GXM4 A0A1S4GWY5 A0A1S4J236 A0A084WD80 A0A0N0U6W4 A0A182KA85 A0A182W7E6 Q7Q3R5 A0A182NTM5 A0A1S4EZX7 A0A182HV10 A0A2S2NY27 X1XCS2 A0A1D2N5X5 E9H8B9 A0A0N8CEL9 A0A0P5LVB8 A0A224YWX5 A0A131YI29 A0A182XZX4 A0A293MXU7 A0A087TCA0 A0A0P5A281 A0A0P5E814 A0A0P5HRR2 A0A0P5SS50 A0A162TAP3 A0A0P5BVI6 A0A0P5K519 A0A0P5HNG8 A0A182LZS9 A0A1Z5KW83 A0A182QD76 A0A0P5DRK5 A0A0P5YKY0 A0A0P6ED54 A0A0P5Y1W3 A0A0P5VD78 A0A0P5VV19 A0A0P5HXL1 A0A1E1XQI5 A0A023GK87
PDB
4PWN
E-value=5.14584e-33,
Score=354
Ontologies
GO
Topology
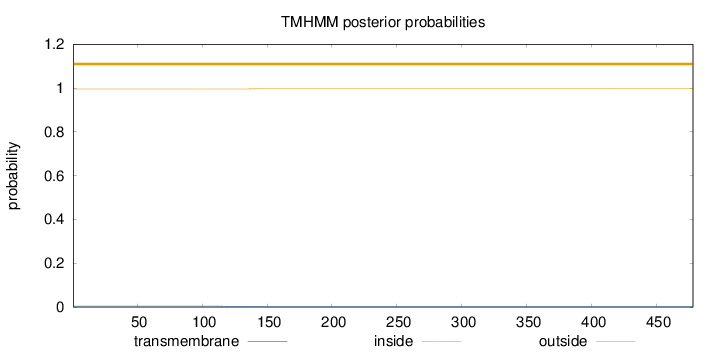
Length:
478
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0356400000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00431
outside
1 - 478
Population Genetic Test Statistics
Pi
207.08941
Theta
185.0536
Tajima's D
0.282489
CLR
156.164953
CSRT
0.445377731113444
Interpretation
Uncertain
