Gene
KWMTBOMO05395
Annotation
PREDICTED:_brahma-associated_protein_of_60_kDa_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.959
Sequence
CDS
ATGCTAACAAGATCAAATTGTCAACAGGCACGCAGCGGATTCGGCGGCGGCGGCGTAGTGGCAGGAGGGGGCGCGCGAGCGGCGGGGGGTGCGGCCTATGGCGGCGGCCCCCGCCCCGCGCCGCCCTCGCCCTCCCCCGTGGCGCCCCCCTCCGCCGTCGCACAGAAGCGCGGGGGTGAGGTCCGCGCCCCGCACCCGCCGCCGCACAAGCCGCTGTACGACGCGTACTCGCAGCCGTCCGCCAAGCGCAAGAAGCGCCTCGCCGACAAGATCCTGCCGCAGAAGGTGCGGGACCTCGTGCCCGAGTCGCAGGCCTACATGGACCTGCTCGCGTTCGAACGCAAACTCGACGCCACCATAATGCGCAAACGTCTCGACATTCAAGAAGCCCTCAAACGCCCCATGAAACAGAAACGAAAACTTCGAATATTCATCTCCAACACGTTCTACCCCGGTCAGGGCGACAACGCGGTGCCGTCGTGGGAGCTGCGCGTGGAGGGCCGTCTGCTCGACGACTCCAAGAACGATCCGAACAAGAATGTCCCGTTGCAGGTGAAGCGGAAGTTTTCGTCGTTCTTCAAGTCTCTGGTGATAGAACTCGATAAAGATCTGTACGGGCCCGACAACCACCTCGTGGAGTGGCACCGCACGGTGACCACGCAGGAGACGGACGGCTTCCAGGTGAAGCGACCCGGCTACAAGAACGTGCGGTGCACCATCCTGCTGCTGCTGGACTACCAGCCGTTGCAGTTCAAGCTGGACCAGCGGCTGGCGCGGCTGTTGGGCGTGCACACGCAGACGCGGCCCGTCATAGTGAACGCGCTGTGGCAGTACGTGAAGACGCACCGCCTGCAGGACCCGCACGAGCGCGAGTACATCGCCTGCGACCGCTACCTCGAGCAGATCTTCGGGGCACCGCGCGTCAAGCTGGCCGAGGTGCCGGCGCGCCTGGGCGCCCTGCTGCACGCGCCCGACCCCATCGTCATCAACCACGTCATCGCCGTGGAGCCGCCGCACGACGCCAAGCAGACCGCCTGCTACGACATCGACGTCGAGGTGGACGACACGCTCAAGGCGCAGATGAACAACTTCCTGCTCAGCACCGCCAACCAGCAGGAGATCCAAGGCCTCGACTCCAAGATCCACGAGACTGTGGACACCATCAACCAGTTGAAAACAAACCGCGAGTTCTTCTTGAGCTTCAGCAAGGATCCGCAGCAGTTCATCCAGAAGTGGCTCGTCAGTCAGTCCAGAGACCTCAAGAGCATGAGCGGCGGCGCCAGCGGCAACCCGGAGGAGGAGCGGCGCGCACAGTTCTACTGCGCGGGCTGGGCGGGCGAGGCGGCCGCGCGACACCTGCACGCGCGCCTGCTGGCCCGCCGCCGCGACCTCGACCTGGCGCTGCGCCCCGCGCTCCTTTAA
Protein
MLTRSNCQQARSGFGGGGVVAGGGARAAGGAAYGGGPRPAPPSPSPVAPPSAVAQKRGGEVRAPHPPPHKPLYDAYSQPSAKRKKRLADKILPQKVRDLVPESQAYMDLLAFERKLDATIMRKRLDIQEALKRPMKQKRKLRIFISNTFYPGQGDNAVPSWELRVEGRLLDDSKNDPNKNVPLQVKRKFSSFFKSLVIELDKDLYGPDNHLVEWHRTVTTQETDGFQVKRPGYKNVRCTILLLLDYQPLQFKLDQRLARLLGVHTQTRPVIVNALWQYVKTHRLQDPHEREYIACDRYLEQIFGAPRVKLAEVPARLGALLHAPDPIVINHVIAVEPPHDAKQTACYDIDVEVDDTLKAQMNNFLLSTANQQEIQGLDSKIHETVDTINQLKTNREFFLSFSKDPQQFIQKWLVSQSRDLKSMSGGASGNPEEERRAQFYCAGWAGEAAARHLHARLLARRRDLDLALRPALL
Summary
Uniprot
A0A212F517
A0A2H1WDF9
S4PE50
A0A232FC06
K7J011
A0A0C9RM20
+ More
A0A158NW78 A0A336K3R2 E9IHD0 A0A195FW01 A0A026VZV2 A0A195EBU8 F4WNG6 A0A195B876 A0A0T6B7F6 A0A151WVH3 N6T5C8 A0A151IQ00 E2BJ35 A0A1W4X460 E2A8V8 D6X1C7 A0A1B6C299 A0A224XB32 A0A154NXR6 J3JT87 A0A310SK33 A0A1B6G9X1 A0A0L7RAZ3 A0A0N0U4U1 A0A1J1IQK7 E0VZ22 U5EWY1 A0A1Y1MM78 A0A0P4VWG3 A0A2A3E676 A0A088ASX9 A0A2J7RJ06 A0A0V0G694 A0A023F5K6 A0A067QVH7 A0A347ZJ96 A0A2S2QL59 A0A2S2PU27 A0A2M4BKN6 J9JYG4 A0A182FHB2 A0A1B0CHL2 W5JNB0 A0A2H8TP99 A0A182N7X6 A0A1Q3F1K6 A0A182VDN5 A0A0K8S4J4 A0A182UBA0 A0A182XD34 Q7Q7F6 A0A0A9WR02 A0A182HMG8 A0A182QTE7 A0A182J057 A0A146M4F7 A0A146ME40 A0A182KD89 A0A182PGF0 A0A0A9X1A5 A0A182YPZ8 A0A084W166 A0A182M0A4 A0A182RX36 A0A1S3DBH4 A0A182SCE2 A0A182WAK3 B0XIC6 A0A1L8DWY3 Q0IEK9 A0A1L8DXP5 A0A1B0DNL8 A0A1S4FN33 A0A182J3D2 T1J8H2 A0A0P4Z628 A0A2J7RIZ4 A0A1A9XGZ4 A0A1B0ANF1 A0A0P6ISZ0 A0A0P5YIF0 A0A0P5AYV7 A0A0N8CBX6 A0A1B0FNR7 A0A0P5ZC21 A0A1A9VRM7 A0A0P5HG74 A0A0P5R174 A0A1A9Z1P4 A0A1A9W9G8 A0A084VVA0 A0A0P4YQM3 A0A0N8A195
A0A158NW78 A0A336K3R2 E9IHD0 A0A195FW01 A0A026VZV2 A0A195EBU8 F4WNG6 A0A195B876 A0A0T6B7F6 A0A151WVH3 N6T5C8 A0A151IQ00 E2BJ35 A0A1W4X460 E2A8V8 D6X1C7 A0A1B6C299 A0A224XB32 A0A154NXR6 J3JT87 A0A310SK33 A0A1B6G9X1 A0A0L7RAZ3 A0A0N0U4U1 A0A1J1IQK7 E0VZ22 U5EWY1 A0A1Y1MM78 A0A0P4VWG3 A0A2A3E676 A0A088ASX9 A0A2J7RJ06 A0A0V0G694 A0A023F5K6 A0A067QVH7 A0A347ZJ96 A0A2S2QL59 A0A2S2PU27 A0A2M4BKN6 J9JYG4 A0A182FHB2 A0A1B0CHL2 W5JNB0 A0A2H8TP99 A0A182N7X6 A0A1Q3F1K6 A0A182VDN5 A0A0K8S4J4 A0A182UBA0 A0A182XD34 Q7Q7F6 A0A0A9WR02 A0A182HMG8 A0A182QTE7 A0A182J057 A0A146M4F7 A0A146ME40 A0A182KD89 A0A182PGF0 A0A0A9X1A5 A0A182YPZ8 A0A084W166 A0A182M0A4 A0A182RX36 A0A1S3DBH4 A0A182SCE2 A0A182WAK3 B0XIC6 A0A1L8DWY3 Q0IEK9 A0A1L8DXP5 A0A1B0DNL8 A0A1S4FN33 A0A182J3D2 T1J8H2 A0A0P4Z628 A0A2J7RIZ4 A0A1A9XGZ4 A0A1B0ANF1 A0A0P6ISZ0 A0A0P5YIF0 A0A0P5AYV7 A0A0N8CBX6 A0A1B0FNR7 A0A0P5ZC21 A0A1A9VRM7 A0A0P5HG74 A0A0P5R174 A0A1A9Z1P4 A0A1A9W9G8 A0A084VVA0 A0A0P4YQM3 A0A0N8A195
Pubmed
EMBL
AGBW02010275
OWR48827.1
ODYU01007913
SOQ51108.1
GAIX01003321
JAA89239.1
+ More
NNAY01000463 OXU28195.1 GBYB01008026 JAG77793.1 ADTU01027831 ADTU01027832 UFQS01000101 UFQT01000101 SSW99606.1 SSX19986.1 GL763242 EFZ20012.1 KQ981215 KYN44606.1 KK107522 QOIP01000001 EZA49333.1 RLU27290.1 KQ979074 KYN22700.1 GL888237 EGI64159.1 KQ976565 KYM80450.1 LJIG01009353 KRT83250.1 KQ982706 KYQ51838.1 APGK01043268 KB741014 KB632328 ENN75399.1 ERL92535.1 KQ976801 KYN08229.1 GL448558 EFN84185.1 GL437663 EFN70187.1 KQ971372 EFA10612.1 GEDC01029660 JAS07638.1 GFTR01006810 JAW09616.1 KQ434781 KZC04466.1 BT126443 AEE61407.1 KQ760498 OAD60089.1 GECZ01010549 JAS59220.1 KQ414618 KOC67995.1 KQ435798 KOX73452.1 CVRI01000054 CRL00785.1 DS235849 EEB18628.1 GANO01001203 JAB58668.1 GEZM01028964 JAV86188.1 GDKW01000135 JAI56460.1 KZ288361 PBC26994.1 NEVH01003011 PNF40806.1 GECL01002613 JAP03511.1 GBBI01002275 JAC16437.1 KK852969 KDR13121.1 FX985664 BBA84402.1 GGMS01009067 MBY78270.1 GGMR01020294 MBY32913.1 GGFJ01004498 MBW53639.1 ABLF02036303 ABLF02036310 AJWK01012454 ADMH02001057 ETN64259.1 GFXV01004034 MBW15839.1 GFDL01013610 JAV21435.1 GBRD01018162 JAG47665.1 AAAB01008960 EAA11403.3 GBHO01032717 GBRD01018161 JAG10887.1 JAG47666.1 APCN01004434 AXCN02000907 AXCP01009255 GDHC01005179 JAQ13450.1 GDHC01000635 JAQ17994.1 GBHO01032719 GBRD01018164 GDHC01006913 GDHC01002473 JAG10885.1 JAG47663.1 JAQ11716.1 JAQ16156.1 ATLV01019212 KE525264 KFB43960.1 AXCM01004189 DS233289 EDS29153.1 GFDF01003180 JAV10904.1 CH477600 EAT38454.1 GFDF01003059 JAV11025.1 AJVK01001415 JH431954 GDIP01218123 JAJ05279.1 PNF40805.1 JXJN01000809 GDIQ01005681 JAN89056.1 GDIP01058997 JAM44718.1 GDIP01192376 GDIQ01120394 JAJ31026.1 JAL31332.1 GDIQ01094851 GDIQ01092700 JAL56875.1 CCAG010016826 GDIP01045810 JAM57905.1 GDIQ01233496 JAK18229.1 GDIQ01109336 JAL42390.1 ATLV01017167 KE525157 KFB41894.1 GDIP01226076 JAI97325.1 GDIP01192377 JAJ31025.1
NNAY01000463 OXU28195.1 GBYB01008026 JAG77793.1 ADTU01027831 ADTU01027832 UFQS01000101 UFQT01000101 SSW99606.1 SSX19986.1 GL763242 EFZ20012.1 KQ981215 KYN44606.1 KK107522 QOIP01000001 EZA49333.1 RLU27290.1 KQ979074 KYN22700.1 GL888237 EGI64159.1 KQ976565 KYM80450.1 LJIG01009353 KRT83250.1 KQ982706 KYQ51838.1 APGK01043268 KB741014 KB632328 ENN75399.1 ERL92535.1 KQ976801 KYN08229.1 GL448558 EFN84185.1 GL437663 EFN70187.1 KQ971372 EFA10612.1 GEDC01029660 JAS07638.1 GFTR01006810 JAW09616.1 KQ434781 KZC04466.1 BT126443 AEE61407.1 KQ760498 OAD60089.1 GECZ01010549 JAS59220.1 KQ414618 KOC67995.1 KQ435798 KOX73452.1 CVRI01000054 CRL00785.1 DS235849 EEB18628.1 GANO01001203 JAB58668.1 GEZM01028964 JAV86188.1 GDKW01000135 JAI56460.1 KZ288361 PBC26994.1 NEVH01003011 PNF40806.1 GECL01002613 JAP03511.1 GBBI01002275 JAC16437.1 KK852969 KDR13121.1 FX985664 BBA84402.1 GGMS01009067 MBY78270.1 GGMR01020294 MBY32913.1 GGFJ01004498 MBW53639.1 ABLF02036303 ABLF02036310 AJWK01012454 ADMH02001057 ETN64259.1 GFXV01004034 MBW15839.1 GFDL01013610 JAV21435.1 GBRD01018162 JAG47665.1 AAAB01008960 EAA11403.3 GBHO01032717 GBRD01018161 JAG10887.1 JAG47666.1 APCN01004434 AXCN02000907 AXCP01009255 GDHC01005179 JAQ13450.1 GDHC01000635 JAQ17994.1 GBHO01032719 GBRD01018164 GDHC01006913 GDHC01002473 JAG10885.1 JAG47663.1 JAQ11716.1 JAQ16156.1 ATLV01019212 KE525264 KFB43960.1 AXCM01004189 DS233289 EDS29153.1 GFDF01003180 JAV10904.1 CH477600 EAT38454.1 GFDF01003059 JAV11025.1 AJVK01001415 JH431954 GDIP01218123 JAJ05279.1 PNF40805.1 JXJN01000809 GDIQ01005681 JAN89056.1 GDIP01058997 JAM44718.1 GDIP01192376 GDIQ01120394 JAJ31026.1 JAL31332.1 GDIQ01094851 GDIQ01092700 JAL56875.1 CCAG010016826 GDIP01045810 JAM57905.1 GDIQ01233496 JAK18229.1 GDIQ01109336 JAL42390.1 ATLV01017167 KE525157 KFB41894.1 GDIP01226076 JAI97325.1 GDIP01192377 JAJ31025.1
Proteomes
UP000007151
UP000215335
UP000002358
UP000005205
UP000078541
UP000053097
+ More
UP000279307 UP000078492 UP000007755 UP000078540 UP000075809 UP000019118 UP000030742 UP000078542 UP000008237 UP000192223 UP000000311 UP000007266 UP000076502 UP000053825 UP000053105 UP000183832 UP000009046 UP000242457 UP000005203 UP000235965 UP000027135 UP000007819 UP000069272 UP000092461 UP000000673 UP000075884 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075886 UP000075880 UP000075881 UP000075885 UP000076408 UP000030765 UP000075883 UP000075900 UP000079169 UP000075901 UP000075920 UP000002320 UP000008820 UP000092462 UP000092443 UP000092460 UP000092444 UP000078200 UP000092445 UP000091820
UP000279307 UP000078492 UP000007755 UP000078540 UP000075809 UP000019118 UP000030742 UP000078542 UP000008237 UP000192223 UP000000311 UP000007266 UP000076502 UP000053825 UP000053105 UP000183832 UP000009046 UP000242457 UP000005203 UP000235965 UP000027135 UP000007819 UP000069272 UP000092461 UP000000673 UP000075884 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075886 UP000075880 UP000075881 UP000075885 UP000076408 UP000030765 UP000075883 UP000075900 UP000079169 UP000075901 UP000075920 UP000002320 UP000008820 UP000092462 UP000092443 UP000092460 UP000092444 UP000078200 UP000092445 UP000091820
Interpro
IPR019835
SWIB_domain
+ More
IPR038041 SMARCD1
IPR003121 SWIB_MDM2_domain
IPR036885 SWIB_MDM2_dom_sf
IPR002553 Clathrin/coatomer_adapt-like_N
IPR013041 Clathrin_app_Ig-like_sf
IPR027417 P-loop_NTPase
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR011989 ARM-like
IPR006689 Small_GTPase_ARF/SAR
IPR008153 GAE_dom
IPR016024 ARM-type_fold
IPR038041 SMARCD1
IPR003121 SWIB_MDM2_domain
IPR036885 SWIB_MDM2_dom_sf
IPR002553 Clathrin/coatomer_adapt-like_N
IPR013041 Clathrin_app_Ig-like_sf
IPR027417 P-loop_NTPase
IPR008152 Clathrin_a/b/g-adaptin_app_Ig
IPR011989 ARM-like
IPR006689 Small_GTPase_ARF/SAR
IPR008153 GAE_dom
IPR016024 ARM-type_fold
Gene 3D
ProteinModelPortal
A0A212F517
A0A2H1WDF9
S4PE50
A0A232FC06
K7J011
A0A0C9RM20
+ More
A0A158NW78 A0A336K3R2 E9IHD0 A0A195FW01 A0A026VZV2 A0A195EBU8 F4WNG6 A0A195B876 A0A0T6B7F6 A0A151WVH3 N6T5C8 A0A151IQ00 E2BJ35 A0A1W4X460 E2A8V8 D6X1C7 A0A1B6C299 A0A224XB32 A0A154NXR6 J3JT87 A0A310SK33 A0A1B6G9X1 A0A0L7RAZ3 A0A0N0U4U1 A0A1J1IQK7 E0VZ22 U5EWY1 A0A1Y1MM78 A0A0P4VWG3 A0A2A3E676 A0A088ASX9 A0A2J7RJ06 A0A0V0G694 A0A023F5K6 A0A067QVH7 A0A347ZJ96 A0A2S2QL59 A0A2S2PU27 A0A2M4BKN6 J9JYG4 A0A182FHB2 A0A1B0CHL2 W5JNB0 A0A2H8TP99 A0A182N7X6 A0A1Q3F1K6 A0A182VDN5 A0A0K8S4J4 A0A182UBA0 A0A182XD34 Q7Q7F6 A0A0A9WR02 A0A182HMG8 A0A182QTE7 A0A182J057 A0A146M4F7 A0A146ME40 A0A182KD89 A0A182PGF0 A0A0A9X1A5 A0A182YPZ8 A0A084W166 A0A182M0A4 A0A182RX36 A0A1S3DBH4 A0A182SCE2 A0A182WAK3 B0XIC6 A0A1L8DWY3 Q0IEK9 A0A1L8DXP5 A0A1B0DNL8 A0A1S4FN33 A0A182J3D2 T1J8H2 A0A0P4Z628 A0A2J7RIZ4 A0A1A9XGZ4 A0A1B0ANF1 A0A0P6ISZ0 A0A0P5YIF0 A0A0P5AYV7 A0A0N8CBX6 A0A1B0FNR7 A0A0P5ZC21 A0A1A9VRM7 A0A0P5HG74 A0A0P5R174 A0A1A9Z1P4 A0A1A9W9G8 A0A084VVA0 A0A0P4YQM3 A0A0N8A195
A0A158NW78 A0A336K3R2 E9IHD0 A0A195FW01 A0A026VZV2 A0A195EBU8 F4WNG6 A0A195B876 A0A0T6B7F6 A0A151WVH3 N6T5C8 A0A151IQ00 E2BJ35 A0A1W4X460 E2A8V8 D6X1C7 A0A1B6C299 A0A224XB32 A0A154NXR6 J3JT87 A0A310SK33 A0A1B6G9X1 A0A0L7RAZ3 A0A0N0U4U1 A0A1J1IQK7 E0VZ22 U5EWY1 A0A1Y1MM78 A0A0P4VWG3 A0A2A3E676 A0A088ASX9 A0A2J7RJ06 A0A0V0G694 A0A023F5K6 A0A067QVH7 A0A347ZJ96 A0A2S2QL59 A0A2S2PU27 A0A2M4BKN6 J9JYG4 A0A182FHB2 A0A1B0CHL2 W5JNB0 A0A2H8TP99 A0A182N7X6 A0A1Q3F1K6 A0A182VDN5 A0A0K8S4J4 A0A182UBA0 A0A182XD34 Q7Q7F6 A0A0A9WR02 A0A182HMG8 A0A182QTE7 A0A182J057 A0A146M4F7 A0A146ME40 A0A182KD89 A0A182PGF0 A0A0A9X1A5 A0A182YPZ8 A0A084W166 A0A182M0A4 A0A182RX36 A0A1S3DBH4 A0A182SCE2 A0A182WAK3 B0XIC6 A0A1L8DWY3 Q0IEK9 A0A1L8DXP5 A0A1B0DNL8 A0A1S4FN33 A0A182J3D2 T1J8H2 A0A0P4Z628 A0A2J7RIZ4 A0A1A9XGZ4 A0A1B0ANF1 A0A0P6ISZ0 A0A0P5YIF0 A0A0P5AYV7 A0A0N8CBX6 A0A1B0FNR7 A0A0P5ZC21 A0A1A9VRM7 A0A0P5HG74 A0A0P5R174 A0A1A9Z1P4 A0A1A9W9G8 A0A084VVA0 A0A0P4YQM3 A0A0N8A195
PDB
1UHR
E-value=1.11361e-27,
Score=307
Ontologies
GO
PANTHER
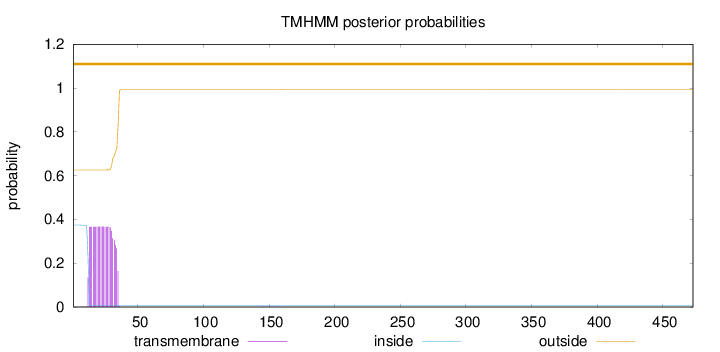
Topology
Subcellular location
Cytoplasm
Length:
473
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.05880999999996
Exp number, first 60 AAs:
8.05458
Total prob of N-in:
0.37409
outside
1 - 473
Population Genetic Test Statistics
Pi
18.668349
Theta
12.509272
Tajima's D
-0.739344
CLR
143.681375
CSRT
0.193790310484476
Interpretation
Uncertain
