Gene
KWMTBOMO05385
Annotation
hypothetical_protein_KGM_01125_[Danaus_plexippus]
Full name
Transmembrane protein 138
Location in the cell
PlasmaMembrane Reliability : 4.827
Sequence
CDS
ATGAAGCTTTCTACGCCAAAATACACGATATGTATAATTATTCAGTTTATTTTCATGTTTTGCGACTTAATATTTAATTGTGTGAGTTTATTTCCGAGAACTAAGGATGCACTTCTTGTATTATTCATATTTCAGGACCTGTTCTTGGTCCTATCTATAACAGCGATGCTGATGACGTTCTTCTCCACCTATCTATTTCAAGCTGGTCTGGTAGAGGTGCTGGTGCGCAAGTTCCGCGCGGCGGGCGCGGTGTGCGCGGCGTACGCGGCGGCCAGCGTGCTGCTGCATGCGGTGTGGCTGCAGCAGCAGTGGCACGAGCCGGGCTCGGCGTCGAGACCAGCGGTCATATTCCTGTTCTTGGTCCAGAGATGCTTGTCCCCCTGGTACTACTTCTTCTACAAACGAGCAGCTCTGCGCGTTAGTGATCCTCGCTTCTATGAAGACATAGAATGGATAAGTCATCAGTTACAGCAACATTGA
Protein
MKLSTPKYTICIIIQFIFMFCDLIFNCVSLFPRTKDALLVLFIFQDLFLVLSITAMLMTFFSTYLFQAGLVEVLVRKFRAAGAVCAAYAAASVLLHAVWLQQQWHEPGSASRPAVIFLFLVQRCLSPWYYFFYKRAALRVSDPRFYEDIEWISHQLQQH
Summary
Description
Required for ciliogenesis.
Miscellaneous
TMEM138 and TMEM216 genes are adjacent and are aligned in a head-to-tail configuration. They share some cis regulatory region and display coordinated expression. Genes were joined by chromosomal rearrangement at the amphiboan to reptile evolutionary transition around 340 million years ago (PubMed:22282472).
Similarity
Belongs to the TMEM138 family.
Keywords
Alternative splicing
Cell projection
Ciliopathy
Cilium
Complete proteome
Disease mutation
Glycoprotein
Joubert syndrome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Vacuole
Feature
chain Transmembrane protein 138
splice variant In isoform 2.
sequence variant In JBTS16; dbSNP:rs387907132.
splice variant In isoform 2.
sequence variant In JBTS16; dbSNP:rs387907132.
Uniprot
A0A2H1WCN1
A0A212EWD6
A0A2W1BHV5
A0A3S2PDB8
A0A0L7LQ38
A0A194QIS6
+ More
A0A0N1I9X7 I4DN49 A0A2A4K8I9 E0VIA7 A0A0T6AWT8 A0A067QR57 A0A084WD31 A7T130 A0A182XB63 A0A182IY92 A0A182VSY7 A0A182R6V6 A0A182K0A6 A0A3M6UNE5 A0A182N8Q3 A0A182P8C8 A0A182R0V3 A0NG40 A0A182I0Q7 A0A2B4S3Z4 A0A1I7SWF8 A0A2M4C270 A0A2M4C1P2 A0A1W4X1S6 A0A2M4CJ02 Q16Z35 A0A023EHC6 A0A1S3JN00 A0A182H3Z8 A0A0K2UPC7 A0A182FMU7 A0A2M4AYE6 T1EB80 A0A2M4AYF8 V3ZVN2 R7VJC8 A0A026WMF2 A0A2P8Y7A3 U4UN81 A0A1Q3FI50 B0XJ62 A0A210PZW4 E2AKA4 B3SC41 F4WLS7 A0A151I3Z3 A0A158NXS8 A0A232EQN6 C3XQZ3 A0A0L7RH53 A0A2G8JFW3 A0A0B2VN96 A0A0M8ZRK5 A0A224XMZ5 T1G6Z5 A0A1U7RYB7 R4FKZ8 A0A1U7SL42 H2ND83 A0A0B1T1Z0 A0A182SKX4 A0A151NXQ0 G5AYG3 A0A1S3G6W0 S4RZE2 A0A3Q0DFM5 A0A3Q2DCL1 G3QN90 A0A3S1BDN2 A0A2R8Z879 H2Q3U1 A0A1B6JFJ2 H0XEQ1 A0A091DGN5 A0A3Q2Q120 T2M540 A0A0L8HUC5 F7HCV4 A0A3Q4HPM5 A0A195FXD2 A0A3Q2PYT9 H9Z9V8 A0A1S3G786 Q9NPI0 A0A3Q2ZKF4 A0A1I8G130 A0A1A6FXW2 A0A2K5HMV8 A0A267DTB1 A0A1A8R4X4 A0A1A8MRQ7 A0A1A8A9S9 A0A1A8BSI4
A0A0N1I9X7 I4DN49 A0A2A4K8I9 E0VIA7 A0A0T6AWT8 A0A067QR57 A0A084WD31 A7T130 A0A182XB63 A0A182IY92 A0A182VSY7 A0A182R6V6 A0A182K0A6 A0A3M6UNE5 A0A182N8Q3 A0A182P8C8 A0A182R0V3 A0NG40 A0A182I0Q7 A0A2B4S3Z4 A0A1I7SWF8 A0A2M4C270 A0A2M4C1P2 A0A1W4X1S6 A0A2M4CJ02 Q16Z35 A0A023EHC6 A0A1S3JN00 A0A182H3Z8 A0A0K2UPC7 A0A182FMU7 A0A2M4AYE6 T1EB80 A0A2M4AYF8 V3ZVN2 R7VJC8 A0A026WMF2 A0A2P8Y7A3 U4UN81 A0A1Q3FI50 B0XJ62 A0A210PZW4 E2AKA4 B3SC41 F4WLS7 A0A151I3Z3 A0A158NXS8 A0A232EQN6 C3XQZ3 A0A0L7RH53 A0A2G8JFW3 A0A0B2VN96 A0A0M8ZRK5 A0A224XMZ5 T1G6Z5 A0A1U7RYB7 R4FKZ8 A0A1U7SL42 H2ND83 A0A0B1T1Z0 A0A182SKX4 A0A151NXQ0 G5AYG3 A0A1S3G6W0 S4RZE2 A0A3Q0DFM5 A0A3Q2DCL1 G3QN90 A0A3S1BDN2 A0A2R8Z879 H2Q3U1 A0A1B6JFJ2 H0XEQ1 A0A091DGN5 A0A3Q2Q120 T2M540 A0A0L8HUC5 F7HCV4 A0A3Q4HPM5 A0A195FXD2 A0A3Q2PYT9 H9Z9V8 A0A1S3G786 Q9NPI0 A0A3Q2ZKF4 A0A1I8G130 A0A1A6FXW2 A0A2K5HMV8 A0A267DTB1 A0A1A8R4X4 A0A1A8MRQ7 A0A1A8A9S9 A0A1A8BSI4
Pubmed
22118469
28756777
26227816
26354079
22651552
20566863
+ More
24845553 24438588 17615350 30382153 12364791 14747013 17210077 21909270 17510324 24945155 26383154 26483478 23254933 24508170 30249741 29403074 23537049 28812685 20798317 18719581 21719571 21347285 28648823 18563158 29023486 22293439 21993625 22398555 22722832 16136131 24065732 25243066 25186727 25319552 14702039 11042152 17974005 16554811 15489334 22282472 26392545
24845553 24438588 17615350 30382153 12364791 14747013 17210077 21909270 17510324 24945155 26383154 26483478 23254933 24508170 30249741 29403074 23537049 28812685 20798317 18719581 21719571 21347285 28648823 18563158 29023486 22293439 21993625 22398555 22722832 16136131 24065732 25243066 25186727 25319552 14702039 11042152 17974005 16554811 15489334 22282472 26392545
EMBL
ODYU01007770
SOQ50823.1
AGBW02012018
OWR45793.1
KZ150352
PZC71253.1
+ More
RSAL01000089 RVE48121.1 JTDY01000359 KOB77552.1 KQ458714 KPJ05417.1 KQ460036 KPJ18443.1 AK402717 BAM19339.1 NWSH01000037 PCG80389.1 DS235184 EEB13113.1 LJIG01022681 KRT79278.1 KK853056 KDR11936.1 ATLV01022925 KE525338 KFB48125.1 DS470072 EDO30334.1 RCHS01001119 RMX55141.1 AXCN02001858 AAAB01008980 EAU76134.1 APCN01005234 LSMT01000171 PFX24621.1 GGFJ01010110 MBW59251.1 GGFJ01010074 MBW59215.1 GGFL01001148 MBW65326.1 CH477499 EAT39915.1 GAPW01005312 JAC08286.1 JXUM01025946 KQ560739 KXJ81063.1 HACA01022549 CDW39910.1 GGFK01012479 MBW45800.1 GAMD01000163 JAB01428.1 GGFK01012506 MBW45827.1 KB203357 ESO84991.1 AMQN01000514 KB291799 ELU18754.1 KK107151 QOIP01000003 EZA57247.1 RLU24393.1 PYGN01000839 PSN40138.1 KB632375 ERL93918.1 GFDL01007836 JAV27209.1 DS233443 EDS30025.1 NEDP02005322 OWF42043.1 GL440210 EFN66129.1 DS985267 EDV19737.1 GL888216 EGI64809.1 KQ976462 KYM84658.1 ADTU01003493 NNAY01002746 OXU20637.1 GG666456 EEN69588.1 KQ414596 KOC70036.1 MRZV01002115 PIK34641.1 JPKZ01001280 KHN82912.1 KQ435944 KOX68203.1 GFTR01002559 JAW13867.1 AMQM01007296 KB097594 ESN93779.1 ACPB03012702 GAHY01002299 JAA75211.1 ABGA01076010 ABGA01076011 NDHI03003558 PNJ22107.1 KN553739 KHJ89812.1 AKHW03001628 KYO41662.1 JH167515 EHB02074.1 CABD030079455 CABD030079456 CABD030079457 RQTK01000155 RUS85944.1 AJFE02100930 AJFE02100931 AACZ04057297 GABD01008324 NBAG03000297 JAA24776.1 PNI45198.1 GECU01009615 JAS98091.1 AAQR03147862 KN122588 KFO29455.1 HAAD01000934 CDG67166.1 KQ417345 KOF92395.1 GAMT01005361 GAMS01008901 GAMR01002042 GAMQ01001457 GAMP01010703 JAB06500.1 JAB14235.1 JAB31890.1 JAB40394.1 JAB42052.1 KQ981204 KYN44972.1 JU475920 AFH32724.1 AK303219 AF151030 AF151032 AL832879 AP003108 BC005201 LZPO01110809 OBS58761.1 NIVC01003245 NIVC01002277 PAA52416.1 PAA59321.1 HAEH01014696 HAEI01016650 SBS00772.1 HAEF01017995 HAEG01004623 SBR59154.1 HADY01012953 HAEJ01017475 SBP51438.1 HADZ01006218 HAEA01013723 SBP70159.1
RSAL01000089 RVE48121.1 JTDY01000359 KOB77552.1 KQ458714 KPJ05417.1 KQ460036 KPJ18443.1 AK402717 BAM19339.1 NWSH01000037 PCG80389.1 DS235184 EEB13113.1 LJIG01022681 KRT79278.1 KK853056 KDR11936.1 ATLV01022925 KE525338 KFB48125.1 DS470072 EDO30334.1 RCHS01001119 RMX55141.1 AXCN02001858 AAAB01008980 EAU76134.1 APCN01005234 LSMT01000171 PFX24621.1 GGFJ01010110 MBW59251.1 GGFJ01010074 MBW59215.1 GGFL01001148 MBW65326.1 CH477499 EAT39915.1 GAPW01005312 JAC08286.1 JXUM01025946 KQ560739 KXJ81063.1 HACA01022549 CDW39910.1 GGFK01012479 MBW45800.1 GAMD01000163 JAB01428.1 GGFK01012506 MBW45827.1 KB203357 ESO84991.1 AMQN01000514 KB291799 ELU18754.1 KK107151 QOIP01000003 EZA57247.1 RLU24393.1 PYGN01000839 PSN40138.1 KB632375 ERL93918.1 GFDL01007836 JAV27209.1 DS233443 EDS30025.1 NEDP02005322 OWF42043.1 GL440210 EFN66129.1 DS985267 EDV19737.1 GL888216 EGI64809.1 KQ976462 KYM84658.1 ADTU01003493 NNAY01002746 OXU20637.1 GG666456 EEN69588.1 KQ414596 KOC70036.1 MRZV01002115 PIK34641.1 JPKZ01001280 KHN82912.1 KQ435944 KOX68203.1 GFTR01002559 JAW13867.1 AMQM01007296 KB097594 ESN93779.1 ACPB03012702 GAHY01002299 JAA75211.1 ABGA01076010 ABGA01076011 NDHI03003558 PNJ22107.1 KN553739 KHJ89812.1 AKHW03001628 KYO41662.1 JH167515 EHB02074.1 CABD030079455 CABD030079456 CABD030079457 RQTK01000155 RUS85944.1 AJFE02100930 AJFE02100931 AACZ04057297 GABD01008324 NBAG03000297 JAA24776.1 PNI45198.1 GECU01009615 JAS98091.1 AAQR03147862 KN122588 KFO29455.1 HAAD01000934 CDG67166.1 KQ417345 KOF92395.1 GAMT01005361 GAMS01008901 GAMR01002042 GAMQ01001457 GAMP01010703 JAB06500.1 JAB14235.1 JAB31890.1 JAB40394.1 JAB42052.1 KQ981204 KYN44972.1 JU475920 AFH32724.1 AK303219 AF151030 AF151032 AL832879 AP003108 BC005201 LZPO01110809 OBS58761.1 NIVC01003245 NIVC01002277 PAA52416.1 PAA59321.1 HAEH01014696 HAEI01016650 SBS00772.1 HAEF01017995 HAEG01004623 SBR59154.1 HADY01012953 HAEJ01017475 SBP51438.1 HADZ01006218 HAEA01013723 SBP70159.1
Proteomes
UP000007151
UP000283053
UP000037510
UP000053268
UP000053240
UP000218220
+ More
UP000009046 UP000027135 UP000030765 UP000001593 UP000076407 UP000075880 UP000075920 UP000075900 UP000075881 UP000275408 UP000075884 UP000075885 UP000075886 UP000007062 UP000075840 UP000225706 UP000095284 UP000192223 UP000008820 UP000085678 UP000069940 UP000249989 UP000069272 UP000030746 UP000014760 UP000053097 UP000279307 UP000245037 UP000030742 UP000002320 UP000242188 UP000000311 UP000009022 UP000007755 UP000078540 UP000005205 UP000215335 UP000001554 UP000053825 UP000230750 UP000031036 UP000053105 UP000015101 UP000189705 UP000015103 UP000189704 UP000001595 UP000075901 UP000050525 UP000006813 UP000081671 UP000245300 UP000265020 UP000001519 UP000271974 UP000240080 UP000002277 UP000005225 UP000028990 UP000265000 UP000053454 UP000008225 UP000261580 UP000078541 UP000005640 UP000264800 UP000095280 UP000092124 UP000233080 UP000215902
UP000009046 UP000027135 UP000030765 UP000001593 UP000076407 UP000075880 UP000075920 UP000075900 UP000075881 UP000275408 UP000075884 UP000075885 UP000075886 UP000007062 UP000075840 UP000225706 UP000095284 UP000192223 UP000008820 UP000085678 UP000069940 UP000249989 UP000069272 UP000030746 UP000014760 UP000053097 UP000279307 UP000245037 UP000030742 UP000002320 UP000242188 UP000000311 UP000009022 UP000007755 UP000078540 UP000005205 UP000215335 UP000001554 UP000053825 UP000230750 UP000031036 UP000053105 UP000015101 UP000189705 UP000015103 UP000189704 UP000001595 UP000075901 UP000050525 UP000006813 UP000081671 UP000245300 UP000265020 UP000001519 UP000271974 UP000240080 UP000002277 UP000005225 UP000028990 UP000265000 UP000053454 UP000008225 UP000261580 UP000078541 UP000005640 UP000264800 UP000095280 UP000092124 UP000233080 UP000215902
PRIDE
Pfam
PF14935 TMEM138
ProteinModelPortal
A0A2H1WCN1
A0A212EWD6
A0A2W1BHV5
A0A3S2PDB8
A0A0L7LQ38
A0A194QIS6
+ More
A0A0N1I9X7 I4DN49 A0A2A4K8I9 E0VIA7 A0A0T6AWT8 A0A067QR57 A0A084WD31 A7T130 A0A182XB63 A0A182IY92 A0A182VSY7 A0A182R6V6 A0A182K0A6 A0A3M6UNE5 A0A182N8Q3 A0A182P8C8 A0A182R0V3 A0NG40 A0A182I0Q7 A0A2B4S3Z4 A0A1I7SWF8 A0A2M4C270 A0A2M4C1P2 A0A1W4X1S6 A0A2M4CJ02 Q16Z35 A0A023EHC6 A0A1S3JN00 A0A182H3Z8 A0A0K2UPC7 A0A182FMU7 A0A2M4AYE6 T1EB80 A0A2M4AYF8 V3ZVN2 R7VJC8 A0A026WMF2 A0A2P8Y7A3 U4UN81 A0A1Q3FI50 B0XJ62 A0A210PZW4 E2AKA4 B3SC41 F4WLS7 A0A151I3Z3 A0A158NXS8 A0A232EQN6 C3XQZ3 A0A0L7RH53 A0A2G8JFW3 A0A0B2VN96 A0A0M8ZRK5 A0A224XMZ5 T1G6Z5 A0A1U7RYB7 R4FKZ8 A0A1U7SL42 H2ND83 A0A0B1T1Z0 A0A182SKX4 A0A151NXQ0 G5AYG3 A0A1S3G6W0 S4RZE2 A0A3Q0DFM5 A0A3Q2DCL1 G3QN90 A0A3S1BDN2 A0A2R8Z879 H2Q3U1 A0A1B6JFJ2 H0XEQ1 A0A091DGN5 A0A3Q2Q120 T2M540 A0A0L8HUC5 F7HCV4 A0A3Q4HPM5 A0A195FXD2 A0A3Q2PYT9 H9Z9V8 A0A1S3G786 Q9NPI0 A0A3Q2ZKF4 A0A1I8G130 A0A1A6FXW2 A0A2K5HMV8 A0A267DTB1 A0A1A8R4X4 A0A1A8MRQ7 A0A1A8A9S9 A0A1A8BSI4
A0A0N1I9X7 I4DN49 A0A2A4K8I9 E0VIA7 A0A0T6AWT8 A0A067QR57 A0A084WD31 A7T130 A0A182XB63 A0A182IY92 A0A182VSY7 A0A182R6V6 A0A182K0A6 A0A3M6UNE5 A0A182N8Q3 A0A182P8C8 A0A182R0V3 A0NG40 A0A182I0Q7 A0A2B4S3Z4 A0A1I7SWF8 A0A2M4C270 A0A2M4C1P2 A0A1W4X1S6 A0A2M4CJ02 Q16Z35 A0A023EHC6 A0A1S3JN00 A0A182H3Z8 A0A0K2UPC7 A0A182FMU7 A0A2M4AYE6 T1EB80 A0A2M4AYF8 V3ZVN2 R7VJC8 A0A026WMF2 A0A2P8Y7A3 U4UN81 A0A1Q3FI50 B0XJ62 A0A210PZW4 E2AKA4 B3SC41 F4WLS7 A0A151I3Z3 A0A158NXS8 A0A232EQN6 C3XQZ3 A0A0L7RH53 A0A2G8JFW3 A0A0B2VN96 A0A0M8ZRK5 A0A224XMZ5 T1G6Z5 A0A1U7RYB7 R4FKZ8 A0A1U7SL42 H2ND83 A0A0B1T1Z0 A0A182SKX4 A0A151NXQ0 G5AYG3 A0A1S3G6W0 S4RZE2 A0A3Q0DFM5 A0A3Q2DCL1 G3QN90 A0A3S1BDN2 A0A2R8Z879 H2Q3U1 A0A1B6JFJ2 H0XEQ1 A0A091DGN5 A0A3Q2Q120 T2M540 A0A0L8HUC5 F7HCV4 A0A3Q4HPM5 A0A195FXD2 A0A3Q2PYT9 H9Z9V8 A0A1S3G786 Q9NPI0 A0A3Q2ZKF4 A0A1I8G130 A0A1A6FXW2 A0A2K5HMV8 A0A267DTB1 A0A1A8R4X4 A0A1A8MRQ7 A0A1A8A9S9 A0A1A8BSI4
Ontologies
PANTHER
Topology
Subcellular location
Vacuole membrane
Cell projection
Cilium
Cell projection
Cilium
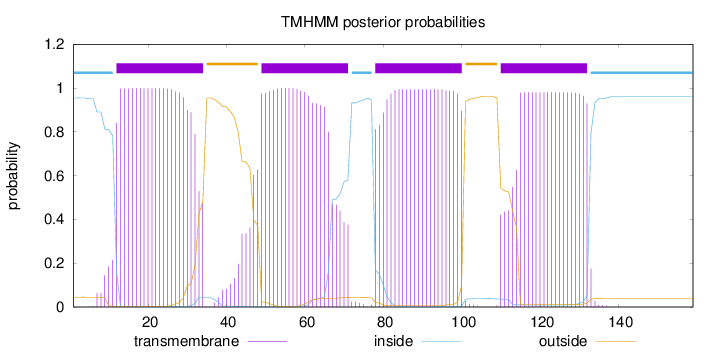
Length:
159
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
87.03763
Exp number, first 60 AAs:
36.80432
Total prob of N-in:
0.95492
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 48
TMhelix
49 - 71
inside
72 - 77
TMhelix
78 - 100
outside
101 - 109
TMhelix
110 - 132
inside
133 - 159
Population Genetic Test Statistics
Pi
164.541968
Theta
153.067167
Tajima's D
0.316887
CLR
0.026576
CSRT
0.462876856157192
Interpretation
Uncertain
