Gene
KWMTBOMO05377 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002549
Annotation
cuticular_protein_RR-1_motif_5_precursor_[Bombyx_mori]
Full name
Flexible cuticle protein 12
Location in the cell
Cytoplasmic Reliability : 1.531 Extracellular Reliability : 1.843
Sequence
CDS
ATGAAATCCTGCATCGTTTTCGCTTTATTCGTGGCCGCTGCCTCGGCCGCCGCCATCACCACCAACAGCAAGGACGCTGTCATCCTTCAACAGACATTTGACAATATCGGCGTTGAGGGTTACGGCTTTGGCTTTGAAACTTCTGATGGCAAAACAGCCCAGGAATCTGCTGTATTGAAGAACGTCGGTACCGAAAATGAGGCTCTTGAGGTTCGCGGACAGTACTCGTACGTCGACCTCGATGGAAAGGTCCACGAGACGACGTACACCGCTGACGAAAATGGTTTCCATCCCTCCGGAGCAGACATTCCCCAGCTCCCTCAAGTCTAA
Protein
MKSCIVFALFVAAASAAAITTNSKDAVILQQTFDNIGVEGYGFGFETSDGKTAQESAVLKNVGTENEALEVRGQYSYVDLDGKVHETTYTADENGFHPSGADIPQLPQV
Summary
Keywords
Cuticle
Signal
Feature
chain Flexible cuticle protein 12
Uniprot
C0H6J6
H9IZ64
Q5MGQ4
D6WMB0
A0A2H1VZY6
A0A2A4J7M4
+ More
B9UCQ9 A0A1J1IRW4 O96052 A0A336LZR1 A0A1L8E3C3 A0A1B0GNX6 A0A336LX25 A0A336LX70 A0A3S2NUQ1 B4IZT2 A0A0R1DW45 S4S7F9 T1DQI3 A0A212EMY2 W5J9B1 A0A1B0ETD6 A0A0T6B9P5 A0A182MR01 Q2LZE0 B4H6H1 P45589 A0A182FCP1 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A182J2Q1 A0A0M3QW63 N6UAA0 I4DLT3 A0A182TZI8 B4ML00 A0A182VC91 B4KWP2 A0A1B0FY22 B0XA35 A0A1J1IV07 Q2TAR4 B3NGE1 A0A1S4FZ80 Q16IX7 B4HUQ2 B4QJA3 B4PJD4 A0A023EEV7 A0A1I8PE08 A0A3B0KAH0 A0A1I8JTU6 A0A182U7E5 Q7Q678 A0A182S4X6 A0A0L7KLZ5 A0A182GLK2 A0A182KPH6 A0A182QGV2 A0A084W0T7 F4WQ42 A0A182V2R6 A0A182SIT2 A0A2A4J8W9 F4WQ43 B3MA22 A0A182IRT6 A0A0L7LB41 Q5XKU2 A0A158NX04 A0A1I8NR37 A0A1I8NP48 A0A1B0CLL0 Q8I0P8 A0A182P5L0 A0A151JU25 A0A1W4UWU2 A0A023EDS8 A0A1Q3FIR8 A0A212EMZ3 B0XA27 A0A182Q833 A0A1L8E379 W5J9A5 A0A0T6B6X8 A0A023EEV6 A0A212FK23 Q16KU4 B4H6G7 A0A0K8VPS1 A0A034WTU9 B4LHE8 A0A151I5M3 A0A182S586 Q16Q11 A0A0K8VZN1 A0A151IHM0 A0A182HF08
B9UCQ9 A0A1J1IRW4 O96052 A0A336LZR1 A0A1L8E3C3 A0A1B0GNX6 A0A336LX25 A0A336LX70 A0A3S2NUQ1 B4IZT2 A0A0R1DW45 S4S7F9 T1DQI3 A0A212EMY2 W5J9B1 A0A1B0ETD6 A0A0T6B9P5 A0A182MR01 Q2LZE0 B4H6H1 P45589 A0A182FCP1 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A182J2Q1 A0A0M3QW63 N6UAA0 I4DLT3 A0A182TZI8 B4ML00 A0A182VC91 B4KWP2 A0A1B0FY22 B0XA35 A0A1J1IV07 Q2TAR4 B3NGE1 A0A1S4FZ80 Q16IX7 B4HUQ2 B4QJA3 B4PJD4 A0A023EEV7 A0A1I8PE08 A0A3B0KAH0 A0A1I8JTU6 A0A182U7E5 Q7Q678 A0A182S4X6 A0A0L7KLZ5 A0A182GLK2 A0A182KPH6 A0A182QGV2 A0A084W0T7 F4WQ42 A0A182V2R6 A0A182SIT2 A0A2A4J8W9 F4WQ43 B3MA22 A0A182IRT6 A0A0L7LB41 Q5XKU2 A0A158NX04 A0A1I8NR37 A0A1I8NP48 A0A1B0CLL0 Q8I0P8 A0A182P5L0 A0A151JU25 A0A1W4UWU2 A0A023EDS8 A0A1Q3FIR8 A0A212EMZ3 B0XA27 A0A182Q833 A0A1L8E379 W5J9A5 A0A0T6B6X8 A0A023EEV6 A0A212FK23 Q16KU4 B4H6G7 A0A0K8VPS1 A0A034WTU9 B4LHE8 A0A151I5M3 A0A182S586 Q16Q11 A0A0K8VZN1 A0A151IHM0 A0A182HF08
Pubmed
19280704
19121390
16023793
18362917
19820115
10327600
+ More
11267900 17994087 17550304 22118469 20920257 23761445 15632085 7535619 20966253 12364791 14747013 17210077 23537049 22651552 17510324 22936249 24945155 26227816 26483478 24438588 21719571 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373
11267900 17994087 17550304 22118469 20920257 23761445 15632085 7535619 20966253 12364791 14747013 17210077 23537049 22651552 17510324 22936249 24945155 26227816 26483478 24438588 21719571 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373
EMBL
BR000506
FAA00507.1
BABH01011198
AY829732
AAV91346.1
KQ971343
+ More
EFA04243.1 ODYU01005497 SOQ46410.1 NWSH01002504 PCG68127.1 EU526836 ACD74812.1 CVRI01000059 CRL02971.1 BABH01011196 AB012081 AB021883 BR000505 BAA36273.1 BAA81902.1 FAA00506.1 UFQT01000259 SSX22511.1 GFDF01000863 JAV13221.1 AJVK01014117 UFQS01000259 SSX02137.1 SSX22514.1 SSX22515.1 RSAL01000175 RVE45046.1 CH916366 EDV95667.1 CM000159 KRK01273.1 JF415079 AFC88812.1 GAMD01001140 JAB00451.1 AGBW02013768 AGBW02008154 OWR42844.1 OWR54092.1 ADMH02002086 ETN59435.1 AJWK01010377 LJIG01002866 KRT84074.1 AXCM01013823 CH379069 EAL29569.1 CH479214 EDW33399.1 U08026 APCN01000091 AAAB01008960 EAA11701.4 CP012525 ALC43212.1 ALC43599.1 APGK01032634 KB740848 KB631977 ENN78645.1 ERL87573.1 AK402251 BAM18873.1 CH963847 EDW72925.1 CH933809 EDW17489.1 AJVK01014120 DS232561 EDS43461.1 CRL02974.1 BC110760 AAI10761.1 CH954178 EDV51107.1 CH478046 EAT34219.1 CH480817 EDW50673.1 CM000363 CM002912 EDX09412.1 KMY97872.1 EDW93602.1 GAPW01006364 JAC07234.1 OUUW01000009 SPP85120.1 EAA11745.1 JTDY01009096 KOB64106.1 JXUM01036286 JXUM01072280 KQ562712 KQ561090 KXJ75275.1 KXJ79736.1 AXCN02000482 ATLV01019159 KE525263 KFB43831.1 GL888262 EGI63665.1 PCG68128.1 EGI63666.1 CH902618 EDV41242.1 JTDY01001889 KOB72625.1 AY769984 AAU94927.1 ADTU01002988 AJWK01017420 AE014296 BT001606 KX532120 AAN12088.1 AAN71361.1 ANY27930.1 KQ981799 KYN35686.1 GAPW01005855 JAC07743.1 GFDL01007568 JAV27477.1 OWR42846.1 EDS43453.1 GFDF01000891 JAV13193.1 ETN59430.1 LJIG01009416 KRT83124.1 GAPW01006363 JAC07235.1 OWR54093.1 CH477938 EAT34914.1 EDW33395.1 EDW33397.1 GDHF01011430 JAI40884.1 GAKP01001205 JAC57747.1 CH940647 EDW68478.1 KQ976434 KYM87206.1 CH477766 EAT36465.1 GDHF01007977 JAI44337.1 KQ977586 KYN01677.1 JXUM01036299 KXJ79741.1
EFA04243.1 ODYU01005497 SOQ46410.1 NWSH01002504 PCG68127.1 EU526836 ACD74812.1 CVRI01000059 CRL02971.1 BABH01011196 AB012081 AB021883 BR000505 BAA36273.1 BAA81902.1 FAA00506.1 UFQT01000259 SSX22511.1 GFDF01000863 JAV13221.1 AJVK01014117 UFQS01000259 SSX02137.1 SSX22514.1 SSX22515.1 RSAL01000175 RVE45046.1 CH916366 EDV95667.1 CM000159 KRK01273.1 JF415079 AFC88812.1 GAMD01001140 JAB00451.1 AGBW02013768 AGBW02008154 OWR42844.1 OWR54092.1 ADMH02002086 ETN59435.1 AJWK01010377 LJIG01002866 KRT84074.1 AXCM01013823 CH379069 EAL29569.1 CH479214 EDW33399.1 U08026 APCN01000091 AAAB01008960 EAA11701.4 CP012525 ALC43212.1 ALC43599.1 APGK01032634 KB740848 KB631977 ENN78645.1 ERL87573.1 AK402251 BAM18873.1 CH963847 EDW72925.1 CH933809 EDW17489.1 AJVK01014120 DS232561 EDS43461.1 CRL02974.1 BC110760 AAI10761.1 CH954178 EDV51107.1 CH478046 EAT34219.1 CH480817 EDW50673.1 CM000363 CM002912 EDX09412.1 KMY97872.1 EDW93602.1 GAPW01006364 JAC07234.1 OUUW01000009 SPP85120.1 EAA11745.1 JTDY01009096 KOB64106.1 JXUM01036286 JXUM01072280 KQ562712 KQ561090 KXJ75275.1 KXJ79736.1 AXCN02000482 ATLV01019159 KE525263 KFB43831.1 GL888262 EGI63665.1 PCG68128.1 EGI63666.1 CH902618 EDV41242.1 JTDY01001889 KOB72625.1 AY769984 AAU94927.1 ADTU01002988 AJWK01017420 AE014296 BT001606 KX532120 AAN12088.1 AAN71361.1 ANY27930.1 KQ981799 KYN35686.1 GAPW01005855 JAC07743.1 GFDL01007568 JAV27477.1 OWR42846.1 EDS43453.1 GFDF01000891 JAV13193.1 ETN59430.1 LJIG01009416 KRT83124.1 GAPW01006363 JAC07235.1 OWR54093.1 CH477938 EAT34914.1 EDW33395.1 EDW33397.1 GDHF01011430 JAI40884.1 GAKP01001205 JAC57747.1 CH940647 EDW68478.1 KQ976434 KYM87206.1 CH477766 EAT36465.1 GDHF01007977 JAI44337.1 KQ977586 KYN01677.1 JXUM01036299 KXJ79741.1
Proteomes
UP000005204
UP000007266
UP000218220
UP000183832
UP000092462
UP000283053
+ More
UP000001070 UP000002282 UP000007151 UP000000673 UP000092461 UP000075883 UP000001819 UP000008744 UP000069272 UP000075840 UP000075882 UP000007062 UP000075880 UP000092553 UP000019118 UP000030742 UP000075902 UP000007798 UP000075903 UP000009192 UP000002320 UP000008711 UP000008820 UP000001292 UP000000304 UP000095300 UP000268350 UP000075900 UP000037510 UP000069940 UP000249989 UP000075886 UP000030765 UP000007755 UP000075901 UP000007801 UP000005205 UP000000803 UP000075885 UP000078541 UP000192221 UP000008792 UP000078540 UP000078542
UP000001070 UP000002282 UP000007151 UP000000673 UP000092461 UP000075883 UP000001819 UP000008744 UP000069272 UP000075840 UP000075882 UP000007062 UP000075880 UP000092553 UP000019118 UP000030742 UP000075902 UP000007798 UP000075903 UP000009192 UP000002320 UP000008711 UP000008820 UP000001292 UP000000304 UP000095300 UP000268350 UP000075900 UP000037510 UP000069940 UP000249989 UP000075886 UP000030765 UP000007755 UP000075901 UP000007801 UP000005205 UP000000803 UP000075885 UP000078541 UP000192221 UP000008792 UP000078540 UP000078542
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6J6
H9IZ64
Q5MGQ4
D6WMB0
A0A2H1VZY6
A0A2A4J7M4
+ More
B9UCQ9 A0A1J1IRW4 O96052 A0A336LZR1 A0A1L8E3C3 A0A1B0GNX6 A0A336LX25 A0A336LX70 A0A3S2NUQ1 B4IZT2 A0A0R1DW45 S4S7F9 T1DQI3 A0A212EMY2 W5J9B1 A0A1B0ETD6 A0A0T6B9P5 A0A182MR01 Q2LZE0 B4H6H1 P45589 A0A182FCP1 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A182J2Q1 A0A0M3QW63 N6UAA0 I4DLT3 A0A182TZI8 B4ML00 A0A182VC91 B4KWP2 A0A1B0FY22 B0XA35 A0A1J1IV07 Q2TAR4 B3NGE1 A0A1S4FZ80 Q16IX7 B4HUQ2 B4QJA3 B4PJD4 A0A023EEV7 A0A1I8PE08 A0A3B0KAH0 A0A1I8JTU6 A0A182U7E5 Q7Q678 A0A182S4X6 A0A0L7KLZ5 A0A182GLK2 A0A182KPH6 A0A182QGV2 A0A084W0T7 F4WQ42 A0A182V2R6 A0A182SIT2 A0A2A4J8W9 F4WQ43 B3MA22 A0A182IRT6 A0A0L7LB41 Q5XKU2 A0A158NX04 A0A1I8NR37 A0A1I8NP48 A0A1B0CLL0 Q8I0P8 A0A182P5L0 A0A151JU25 A0A1W4UWU2 A0A023EDS8 A0A1Q3FIR8 A0A212EMZ3 B0XA27 A0A182Q833 A0A1L8E379 W5J9A5 A0A0T6B6X8 A0A023EEV6 A0A212FK23 Q16KU4 B4H6G7 A0A0K8VPS1 A0A034WTU9 B4LHE8 A0A151I5M3 A0A182S586 Q16Q11 A0A0K8VZN1 A0A151IHM0 A0A182HF08
B9UCQ9 A0A1J1IRW4 O96052 A0A336LZR1 A0A1L8E3C3 A0A1B0GNX6 A0A336LX25 A0A336LX70 A0A3S2NUQ1 B4IZT2 A0A0R1DW45 S4S7F9 T1DQI3 A0A212EMY2 W5J9B1 A0A1B0ETD6 A0A0T6B9P5 A0A182MR01 Q2LZE0 B4H6H1 P45589 A0A182FCP1 A0A1I8JTT8 A0A182KPF5 Q7PNS3 A0A182J2Q1 A0A0M3QW63 N6UAA0 I4DLT3 A0A182TZI8 B4ML00 A0A182VC91 B4KWP2 A0A1B0FY22 B0XA35 A0A1J1IV07 Q2TAR4 B3NGE1 A0A1S4FZ80 Q16IX7 B4HUQ2 B4QJA3 B4PJD4 A0A023EEV7 A0A1I8PE08 A0A3B0KAH0 A0A1I8JTU6 A0A182U7E5 Q7Q678 A0A182S4X6 A0A0L7KLZ5 A0A182GLK2 A0A182KPH6 A0A182QGV2 A0A084W0T7 F4WQ42 A0A182V2R6 A0A182SIT2 A0A2A4J8W9 F4WQ43 B3MA22 A0A182IRT6 A0A0L7LB41 Q5XKU2 A0A158NX04 A0A1I8NR37 A0A1I8NP48 A0A1B0CLL0 Q8I0P8 A0A182P5L0 A0A151JU25 A0A1W4UWU2 A0A023EDS8 A0A1Q3FIR8 A0A212EMZ3 B0XA27 A0A182Q833 A0A1L8E379 W5J9A5 A0A0T6B6X8 A0A023EEV6 A0A212FK23 Q16KU4 B4H6G7 A0A0K8VPS1 A0A034WTU9 B4LHE8 A0A151I5M3 A0A182S586 Q16Q11 A0A0K8VZN1 A0A151IHM0 A0A182HF08
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.996274
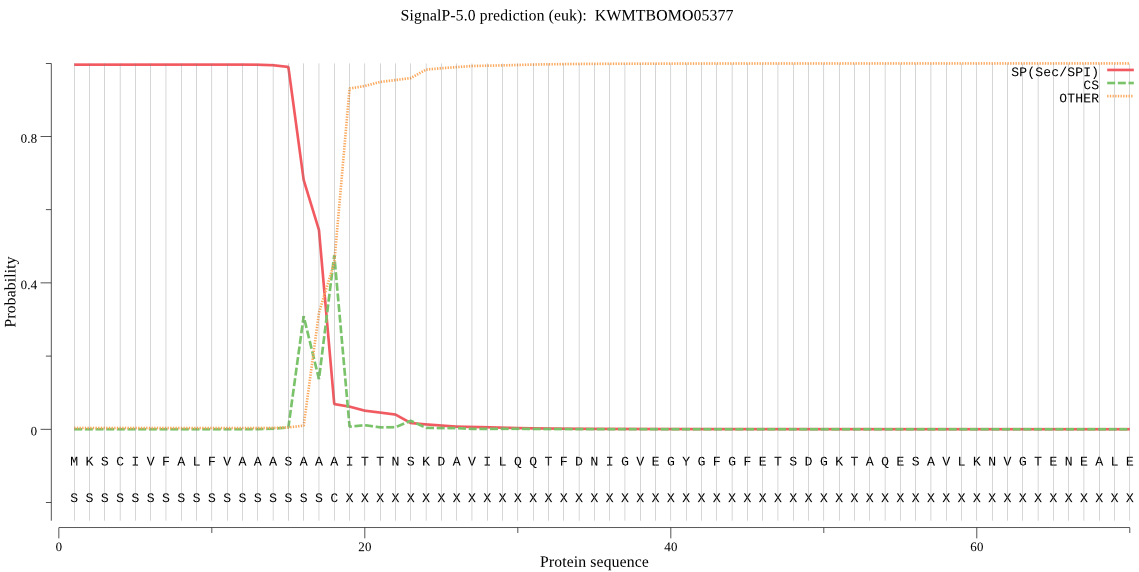
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.96382
Exp number, first 60 AAs:
5.96382
Total prob of N-in:
0.25935
outside
1 - 109

Population Genetic Test Statistics
Pi
192.652142
Theta
158.942107
Tajima's D
0.829121
CLR
0.114213
CSRT
0.617019149042548
Interpretation
Uncertain
