Pre Gene Modal
BGIBMGA002547
Annotation
PREDICTED:_neurexin-4_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.155 Extracellular Reliability : 1.273 PlasmaMembrane Reliability : 1.141
Sequence
CDS
ATGTCTACGTCTCTGTCTGTGGGCAGTCTTCTGGATGACAATATTTGGCACGACGTAGTCATATCTAGGAACAGAAGGGACATCATCTTCTCTGTGGACCGGGTTATAGTGCAGGAGAGAATCAAGGGAGAATTCTCCAGGCTCAACTTGAACAGAGCTCTGTACATAGGGGGCGTGCCGAATTTACAAGAGGGTCTAGTTGTGAACCAAAATTTCACAGGCTGCATAGAGAACATGTACCTCAACGCGACCAACGTTATACAGGATTTGAAAAAGGGCTATGAGAGTGGCGAACCATTTAAGTTCCAGAAACTGAACACCCTGTACGCGTGTCCGGAGCCCCCTGTGGTGCCGGTGACGTTCCTGAAGGAAGGCTCGTACGCCAAGCTGCGCGGGTACTCCGGCGGCAGCGCGCTCAACGTGTCGCTGGAGTTCAGGACCTACGAGCTGCAGGGGCTGCTCGTGTACCACCGGTTCAACACCGAGGGATATGTTAAGGTATATCTTGAAGATGGTAAAGTGAAAGTCGAGTTGGAGACGGCCGGATCTCCTCGCGTGAAGCTGGACAATTACGTGGAGCTGTGGAACGACGGTCGCTGGCACTCGCTGATGCTGACCCTGGCCCCCGACTCGCTGGTGCTGTCCATGGACTCCAGAGCCGTGCACACGTCAAAGAGACTGCGATTTCTTACCGGCAGCTATTATTTTATAGCTGGAGGCAAAGCCCCACCGCGCGGCTTCATCGGTTGCATGCGCAAGATTTCCGTGGACGGGAACTACCGCCTGCCGACGGACTGGAAGAAGGAGGACTACTGCTGCCCCAACGAGCTCGTGTTCGACGCCTGCCAGATGATCGACAGGTGCAGCCCCAACCCCTGCGAGCACGGCGGCCGCTGCGCGCAGACCGCCGACGAGTTCTCCTGCAACTGTCGCGACACGGGCTACGCCGGCGCCGTGTGCCACACGTCGGTGCACCCGCTGTCGTGCCAGGCGTACCACAGCGTGTCGGCGGCCCGGCGCTCCGCCAGCGTGCACATCGACGTGGACGGCTCCGGCCCGCTGCCGCCCTTCCCCGTCACCTGCGAGTTCTACTCGGACGGCCGCGTGATAACGTCGGTGAAGCACTCGGTGTCGCAGAGCACGGTGGTGGACGGCTACCAGGAGGCGGGCAGCTTCCGGCAGGACGTCATCTACGACGCCAACCGGGCGCAGCTCGAGGCCCTGCTCAACAGATCTGTGTCCTGCTCGCAGCGACTGGACTACTGGTGCAGGCACTCGAGGCTCATGAACTCACCTAGCGAGGAGTCCAACTTCCAGCCGTTCGCGTGGTGGGTGTCCCGCTCCGGCCAGCGCATGGACTACTGGGCCGGCGCCACGCCCGGCTCGCGCATGTGCGCCTGCGGCCTGCTGGGCTCCTGTCTCGACAACACCAAGTGGTGCAACTGCGACGCAGAGTACGCGCAGCTGCGCCCCGAGGAGTTCCAGTCGGACGGCGGCGACATCACGGAGAAGGAGTTCCTGCCGGTGAAGCAGCTGCGCTTCGGCGACACTGGCAGCCACCTCGACGAGAAGGAGGGCCGCTACACGCTCGGGCCGCTGCTGTGCGAGGGGGACGATTTGTTCAACAACGCCATCACGTTCCGCGTGGCCGACGCCATCATCCTGCTGCCGACGTTCGACTTGGGGCACAGTGGCGACATCTACTTCGAGTTCAAGACGACCAAGGAGAATGCTGTTCTGCTGCATTCCAAGGGCCCGCAAGACTACATAAAGCTGTCAATAATCGGAGGGGATCAACTGCAGCTGCAGCTGCAAGTGGGTGACACGCCTCTAGGGGTATCCGTGGAGACCAGCAACCGTCTGGCTGACAACCAGTGGCACTCCGTGTCCATCGAAAGGAATAGGAAGGAGGCTAGGCTGGTGGTCGACGGAGCCTTGAAGAACGAGATCCGCACCGTGGGCGGCACGGCGCGCGCCCTGCAGCTGTCCACAGGCCTCGCCCTGGGCGCCGCGCTGGACCGCAGCGACGGGCTGGTGGGCTGCCTGCGCGCCCTGCTGCTCAACGGCAGGCCCGTGCACCTGGCGCCGCTCGCCCGCACCGGGCTGTACGGCATATCCGAGGGCTGCGTCGGCAAGTGCTCGTCGTCGCCGTGTCTCAACAACGGCACGTGCATCGAGGGCTTCGACTCCTACACGTGCGACTGTCGCTGGACGGCCTTCAAAGGCCCCATCTGTGCGGACGAAATCGGCGTAAATCTCCGACCGAACTCGTTGATCAAGTACGATTTCCTGGGCTCGTGGCGCTCCACCATCGCTGAGAAGATCCGGGTCGGCTTCACCACCACCAACCCGAAGGGATTCCTGCTCGGCTTCTACTCGAACATTTCAGGAGAGTACTTGACTCTGATGGTCTCCAATTCCGGTCACCTGCGCGTCGTGTTCGACTTCGGCTTCGAGCGGCAGGAGATCGTGTTCCAGGGGAAGCACTTCGCGCTGGGCCAGTACCACGACGTGCGGCTAGCGCGGCGGGACAGCGGCGCCACGCTCGCGCTGCAGGTGGACAGCTACGAGACGCAGGAGTACCGCTTCAACATCAAGGAGTCGGCGGACGCGCAGTTCAACAACATCCAGTACATGTACATCGGCCGCAACGAGTCCATGGGCGAGGGCTTCGTGGGCTGCGTGAGCCGCGTGGAGTTCGACGACATCTACCCGCTCAAGCTGCTCTTCCAGCAGGACCCGCCGCCCAACGTCAGGAGCCTCGGAGGAGGAGTTTTGCATGAGGACTTCTGCGGCGTGGAGCCCGTGACGCACCCGCCCGAGCCCGTGGAGACCCGGCCGCCGCCCGCCCTCGACCTGCAGCACGAGATCAACTTCCGGAAGACGGACGAAGCCATACTCGGAACGGTGCTGGGGTTCATCTTCCTGCTGCTGGTGGTGGTGGCGGTGGTGCTGGTGCGCGCGCTGTCGCGGCACAAGGGCGAGTACCTGACGCAGGAGGAGCGCGGCGCGGACGGGGCGGCCGACCCCGACGCCGCCGCGCTGGCCGCCGCCACCGGACCGCGCGTCACCAAGCGCAGGGAGTTCTTCATCTAA
Protein
MSTSLSVGSLLDDNIWHDVVISRNRRDIIFSVDRVIVQERIKGEFSRLNLNRALYIGGVPNLQEGLVVNQNFTGCIENMYLNATNVIQDLKKGYESGEPFKFQKLNTLYACPEPPVVPVTFLKEGSYAKLRGYSGGSALNVSLEFRTYELQGLLVYHRFNTEGYVKVYLEDGKVKVELETAGSPRVKLDNYVELWNDGRWHSLMLTLAPDSLVLSMDSRAVHTSKRLRFLTGSYYFIAGGKAPPRGFIGCMRKISVDGNYRLPTDWKKEDYCCPNELVFDACQMIDRCSPNPCEHGGRCAQTADEFSCNCRDTGYAGAVCHTSVHPLSCQAYHSVSAARRSASVHIDVDGSGPLPPFPVTCEFYSDGRVITSVKHSVSQSTVVDGYQEAGSFRQDVIYDANRAQLEALLNRSVSCSQRLDYWCRHSRLMNSPSEESNFQPFAWWVSRSGQRMDYWAGATPGSRMCACGLLGSCLDNTKWCNCDAEYAQLRPEEFQSDGGDITEKEFLPVKQLRFGDTGSHLDEKEGRYTLGPLLCEGDDLFNNAITFRVADAIILLPTFDLGHSGDIYFEFKTTKENAVLLHSKGPQDYIKLSIIGGDQLQLQLQVGDTPLGVSVETSNRLADNQWHSVSIERNRKEARLVVDGALKNEIRTVGGTARALQLSTGLALGAALDRSDGLVGCLRALLLNGRPVHLAPLARTGLYGISEGCVGKCSSSPCLNNGTCIEGFDSYTCDCRWTAFKGPICADEIGVNLRPNSLIKYDFLGSWRSTIAEKIRVGFTTTNPKGFLLGFYSNISGEYLTLMVSNSGHLRVVFDFGFERQEIVFQGKHFALGQYHDVRLARRDSGATLALQVDSYETQEYRFNIKESADAQFNNIQYMYIGRNESMGEGFVGCVSRVEFDDIYPLKLLFQQDPPPNVRSLGGGVLHEDFCGVEPVTHPPEPVETRPPPALDLQHEINFRKTDEAILGTVLGFIFLLLVVVAVVLVRALSRHKGEYLTQEERGADGAADPDAAALAAATGPRVTKRREFFI
Summary
Uniprot
H9IZ62
A0A2H1WVR4
A0A2A4J9Q9
A0A2A4J838
A0A212FIV0
A0A194QKI5
+ More
A0A310S6B2 A0A2J7PDX5 A0A3L8DKW2 A0A026WLY2 A0A067RN92 A0A1B6CD05 U5EK54 A0A1B6CTQ5 A0A0J7KWH4 E2A9Q1 A0A0M9A284 A0A1Y1LIF2 A0A1Y1LEP8 A0A154PT19 A0A2A3E345 A0A195FRF1 A0A088AIQ0 A0A151X154 A0A1L8DMG5 E9IZP1 A0A0L7R366 A0A151J789 A0A195CBV2 A0A1L8DLY5 A0A158NIQ9 A0A336MI58 A0A195BZ68 A0A158NIR0 D6X3H5 A0A139WB44 T1IDR3 A0A0P4VGR3 F4WDG8 A0A1B0D1T5 A0A224X9Z3 E2BRP7 A0A2P8YX59 A0A0C9R2D9 T1PL43 A0A1I8MBA0 A0A1I8MB95 E0VKA9 A0A1B6GVV5 A0A069DXX4 A0A1W4W2V1 A0A1W4WDG7 A0A1I8QCE6 A0A1I8QCH8 A0A232F4M3 B4KZ44 K7IW44 A0A0Q9XNR7 A0A182X3K6 A0A182JQX7 A0A084WNH7 A0A182V8M3 A0A182U038 A0A182I3G4 A0A182KRI3 B4N3T4 A0A182PH94 A0A0Q9X2H0 Q7PT06 A7URA4 Q17AC6 A0A182XZ17 A0A1B0C5X3 A0A1B0FMI3 A0A0Q9WSU9 B4LEI8 A0A0K8VAM6 A0A0P9C1Y0 B4IWN2 A0A1B0A758 A0A1A9VNB0 A0A034V3M2 B3M7V9 A0A034V7Z4 A0A0R3P3K9 B4GR64 D0QWT1 A0A182JK12 A0A0K8SRD6 B0WW64 A0A1B0BXR4 A0A146LPR9 A0A0A1X7R1 A0A182ML99 Q2M0S2 A0A3B0JRE6 W8BBY5 A0A2R7VN87 W8B277 A0A182NH62
A0A310S6B2 A0A2J7PDX5 A0A3L8DKW2 A0A026WLY2 A0A067RN92 A0A1B6CD05 U5EK54 A0A1B6CTQ5 A0A0J7KWH4 E2A9Q1 A0A0M9A284 A0A1Y1LIF2 A0A1Y1LEP8 A0A154PT19 A0A2A3E345 A0A195FRF1 A0A088AIQ0 A0A151X154 A0A1L8DMG5 E9IZP1 A0A0L7R366 A0A151J789 A0A195CBV2 A0A1L8DLY5 A0A158NIQ9 A0A336MI58 A0A195BZ68 A0A158NIR0 D6X3H5 A0A139WB44 T1IDR3 A0A0P4VGR3 F4WDG8 A0A1B0D1T5 A0A224X9Z3 E2BRP7 A0A2P8YX59 A0A0C9R2D9 T1PL43 A0A1I8MBA0 A0A1I8MB95 E0VKA9 A0A1B6GVV5 A0A069DXX4 A0A1W4W2V1 A0A1W4WDG7 A0A1I8QCE6 A0A1I8QCH8 A0A232F4M3 B4KZ44 K7IW44 A0A0Q9XNR7 A0A182X3K6 A0A182JQX7 A0A084WNH7 A0A182V8M3 A0A182U038 A0A182I3G4 A0A182KRI3 B4N3T4 A0A182PH94 A0A0Q9X2H0 Q7PT06 A7URA4 Q17AC6 A0A182XZ17 A0A1B0C5X3 A0A1B0FMI3 A0A0Q9WSU9 B4LEI8 A0A0K8VAM6 A0A0P9C1Y0 B4IWN2 A0A1B0A758 A0A1A9VNB0 A0A034V3M2 B3M7V9 A0A034V7Z4 A0A0R3P3K9 B4GR64 D0QWT1 A0A182JK12 A0A0K8SRD6 B0WW64 A0A1B0BXR4 A0A146LPR9 A0A0A1X7R1 A0A182ML99 Q2M0S2 A0A3B0JRE6 W8BBY5 A0A2R7VN87 W8B277 A0A182NH62
Pubmed
EMBL
BABH01011183
BABH01011184
BABH01011185
BABH01011186
ODYU01011424
SOQ57153.1
+ More
NWSH01002504 PCG68133.1 PCG68131.1 AGBW02008322 OWR53668.1 KQ458714 KPJ05430.1 KQ768090 OAD53194.1 NEVH01026136 PNF14522.1 QOIP01000007 RLU21057.1 KK107152 EZA57067.1 KK852570 KDR21164.1 GEDC01025980 GEDC01025429 JAS11318.1 JAS11869.1 GANO01001940 JAB57931.1 GEDC01020500 GEDC01003483 JAS16798.1 JAS33815.1 LBMM01002611 KMQ94594.1 GL437918 EFN69853.1 KQ435758 KOX75795.1 GEZM01057720 JAV72120.1 GEZM01057721 JAV72119.1 KQ435207 KZC15059.1 KZ288408 PBC26118.1 KQ981285 KYN43175.1 KQ982603 KYQ53967.1 GFDF01006530 JAV07554.1 GL767232 EFZ13948.1 KQ414663 KOC65330.1 KQ979695 KYN19702.1 KQ978068 KYM97578.1 GFDF01006663 JAV07421.1 ADTU01017067 ADTU01017068 UFQS01001052 UFQT01001052 SSX08727.1 SSX28639.1 KQ976394 KYM93213.1 KQ971374 EEZ97422.2 KYB25140.1 ACPB03002180 GDKW01003089 JAI53506.1 GL888087 EGI67748.1 AJVK01022329 AJVK01022330 GFTR01008702 JAW07724.1 GL449978 EFN81641.1 PYGN01000306 PSN48824.1 GBYB01007007 JAG76774.1 KA649467 KA649669 AFP64096.1 DS235241 EEB13815.1 GECZ01003213 JAS66556.1 GBGD01000149 JAC88740.1 NNAY01000997 OXU25562.1 CH933809 EDW17841.1 KRG05784.1 ATLV01024605 KE525352 KFB51771.1 APCN01000208 CH964095 EDW79289.2 KRF99042.1 AAAB01008807 EAA04731.5 EDO64546.1 CH477336 EAT43235.1 JXJN01026284 CCAG010014702 CH940647 KRF84159.1 EDW69073.2 GDHF01016406 JAI35908.1 CH902618 KPU77657.1 CH916366 EDV96258.1 GAKP01021061 JAC37891.1 EDV38832.2 GAKP01021062 JAC37890.1 CH379069 KRT07708.1 CH479188 EDW40249.1 FJ821035 ACN94773.1 GBRD01009936 JAG55888.1 DS232139 EDS35896.1 JXJN01022303 GDHC01009863 JAQ08766.1 GBXI01007356 JAD06936.1 AXCM01004161 EAL30857.3 OUUW01000002 SPP76269.1 GAMC01019186 JAB87369.1 KK854004 PTY09017.1 GAMC01019184 JAB87371.1
NWSH01002504 PCG68133.1 PCG68131.1 AGBW02008322 OWR53668.1 KQ458714 KPJ05430.1 KQ768090 OAD53194.1 NEVH01026136 PNF14522.1 QOIP01000007 RLU21057.1 KK107152 EZA57067.1 KK852570 KDR21164.1 GEDC01025980 GEDC01025429 JAS11318.1 JAS11869.1 GANO01001940 JAB57931.1 GEDC01020500 GEDC01003483 JAS16798.1 JAS33815.1 LBMM01002611 KMQ94594.1 GL437918 EFN69853.1 KQ435758 KOX75795.1 GEZM01057720 JAV72120.1 GEZM01057721 JAV72119.1 KQ435207 KZC15059.1 KZ288408 PBC26118.1 KQ981285 KYN43175.1 KQ982603 KYQ53967.1 GFDF01006530 JAV07554.1 GL767232 EFZ13948.1 KQ414663 KOC65330.1 KQ979695 KYN19702.1 KQ978068 KYM97578.1 GFDF01006663 JAV07421.1 ADTU01017067 ADTU01017068 UFQS01001052 UFQT01001052 SSX08727.1 SSX28639.1 KQ976394 KYM93213.1 KQ971374 EEZ97422.2 KYB25140.1 ACPB03002180 GDKW01003089 JAI53506.1 GL888087 EGI67748.1 AJVK01022329 AJVK01022330 GFTR01008702 JAW07724.1 GL449978 EFN81641.1 PYGN01000306 PSN48824.1 GBYB01007007 JAG76774.1 KA649467 KA649669 AFP64096.1 DS235241 EEB13815.1 GECZ01003213 JAS66556.1 GBGD01000149 JAC88740.1 NNAY01000997 OXU25562.1 CH933809 EDW17841.1 KRG05784.1 ATLV01024605 KE525352 KFB51771.1 APCN01000208 CH964095 EDW79289.2 KRF99042.1 AAAB01008807 EAA04731.5 EDO64546.1 CH477336 EAT43235.1 JXJN01026284 CCAG010014702 CH940647 KRF84159.1 EDW69073.2 GDHF01016406 JAI35908.1 CH902618 KPU77657.1 CH916366 EDV96258.1 GAKP01021061 JAC37891.1 EDV38832.2 GAKP01021062 JAC37890.1 CH379069 KRT07708.1 CH479188 EDW40249.1 FJ821035 ACN94773.1 GBRD01009936 JAG55888.1 DS232139 EDS35896.1 JXJN01022303 GDHC01009863 JAQ08766.1 GBXI01007356 JAD06936.1 AXCM01004161 EAL30857.3 OUUW01000002 SPP76269.1 GAMC01019186 JAB87369.1 KK854004 PTY09017.1 GAMC01019184 JAB87371.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000235965
UP000279307
+ More
UP000053097 UP000027135 UP000036403 UP000000311 UP000053105 UP000076502 UP000242457 UP000078541 UP000005203 UP000075809 UP000053825 UP000078492 UP000078542 UP000005205 UP000078540 UP000007266 UP000015103 UP000007755 UP000092462 UP000008237 UP000245037 UP000095301 UP000009046 UP000192223 UP000095300 UP000215335 UP000009192 UP000002358 UP000076407 UP000075881 UP000030765 UP000075903 UP000075902 UP000075840 UP000075882 UP000007798 UP000075885 UP000007062 UP000008820 UP000076408 UP000092460 UP000092444 UP000008792 UP000007801 UP000001070 UP000092445 UP000078200 UP000001819 UP000008744 UP000075880 UP000002320 UP000075883 UP000268350 UP000075884
UP000053097 UP000027135 UP000036403 UP000000311 UP000053105 UP000076502 UP000242457 UP000078541 UP000005203 UP000075809 UP000053825 UP000078492 UP000078542 UP000005205 UP000078540 UP000007266 UP000015103 UP000007755 UP000092462 UP000008237 UP000245037 UP000095301 UP000009046 UP000192223 UP000095300 UP000215335 UP000009192 UP000002358 UP000076407 UP000075881 UP000030765 UP000075903 UP000075902 UP000075840 UP000075882 UP000007798 UP000075885 UP000007062 UP000008820 UP000076408 UP000092460 UP000092444 UP000008792 UP000007801 UP000001070 UP000092445 UP000078200 UP000001819 UP000008744 UP000075880 UP000002320 UP000075883 UP000268350 UP000075884
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9IZ62
A0A2H1WVR4
A0A2A4J9Q9
A0A2A4J838
A0A212FIV0
A0A194QKI5
+ More
A0A310S6B2 A0A2J7PDX5 A0A3L8DKW2 A0A026WLY2 A0A067RN92 A0A1B6CD05 U5EK54 A0A1B6CTQ5 A0A0J7KWH4 E2A9Q1 A0A0M9A284 A0A1Y1LIF2 A0A1Y1LEP8 A0A154PT19 A0A2A3E345 A0A195FRF1 A0A088AIQ0 A0A151X154 A0A1L8DMG5 E9IZP1 A0A0L7R366 A0A151J789 A0A195CBV2 A0A1L8DLY5 A0A158NIQ9 A0A336MI58 A0A195BZ68 A0A158NIR0 D6X3H5 A0A139WB44 T1IDR3 A0A0P4VGR3 F4WDG8 A0A1B0D1T5 A0A224X9Z3 E2BRP7 A0A2P8YX59 A0A0C9R2D9 T1PL43 A0A1I8MBA0 A0A1I8MB95 E0VKA9 A0A1B6GVV5 A0A069DXX4 A0A1W4W2V1 A0A1W4WDG7 A0A1I8QCE6 A0A1I8QCH8 A0A232F4M3 B4KZ44 K7IW44 A0A0Q9XNR7 A0A182X3K6 A0A182JQX7 A0A084WNH7 A0A182V8M3 A0A182U038 A0A182I3G4 A0A182KRI3 B4N3T4 A0A182PH94 A0A0Q9X2H0 Q7PT06 A7URA4 Q17AC6 A0A182XZ17 A0A1B0C5X3 A0A1B0FMI3 A0A0Q9WSU9 B4LEI8 A0A0K8VAM6 A0A0P9C1Y0 B4IWN2 A0A1B0A758 A0A1A9VNB0 A0A034V3M2 B3M7V9 A0A034V7Z4 A0A0R3P3K9 B4GR64 D0QWT1 A0A182JK12 A0A0K8SRD6 B0WW64 A0A1B0BXR4 A0A146LPR9 A0A0A1X7R1 A0A182ML99 Q2M0S2 A0A3B0JRE6 W8BBY5 A0A2R7VN87 W8B277 A0A182NH62
A0A310S6B2 A0A2J7PDX5 A0A3L8DKW2 A0A026WLY2 A0A067RN92 A0A1B6CD05 U5EK54 A0A1B6CTQ5 A0A0J7KWH4 E2A9Q1 A0A0M9A284 A0A1Y1LIF2 A0A1Y1LEP8 A0A154PT19 A0A2A3E345 A0A195FRF1 A0A088AIQ0 A0A151X154 A0A1L8DMG5 E9IZP1 A0A0L7R366 A0A151J789 A0A195CBV2 A0A1L8DLY5 A0A158NIQ9 A0A336MI58 A0A195BZ68 A0A158NIR0 D6X3H5 A0A139WB44 T1IDR3 A0A0P4VGR3 F4WDG8 A0A1B0D1T5 A0A224X9Z3 E2BRP7 A0A2P8YX59 A0A0C9R2D9 T1PL43 A0A1I8MBA0 A0A1I8MB95 E0VKA9 A0A1B6GVV5 A0A069DXX4 A0A1W4W2V1 A0A1W4WDG7 A0A1I8QCE6 A0A1I8QCH8 A0A232F4M3 B4KZ44 K7IW44 A0A0Q9XNR7 A0A182X3K6 A0A182JQX7 A0A084WNH7 A0A182V8M3 A0A182U038 A0A182I3G4 A0A182KRI3 B4N3T4 A0A182PH94 A0A0Q9X2H0 Q7PT06 A7URA4 Q17AC6 A0A182XZ17 A0A1B0C5X3 A0A1B0FMI3 A0A0Q9WSU9 B4LEI8 A0A0K8VAM6 A0A0P9C1Y0 B4IWN2 A0A1B0A758 A0A1A9VNB0 A0A034V3M2 B3M7V9 A0A034V7Z4 A0A0R3P3K9 B4GR64 D0QWT1 A0A182JK12 A0A0K8SRD6 B0WW64 A0A1B0BXR4 A0A146LPR9 A0A0A1X7R1 A0A182ML99 Q2M0S2 A0A3B0JRE6 W8BBY5 A0A2R7VN87 W8B277 A0A182NH62
PDB
3POY
E-value=5.75691e-28,
Score=313
Ontologies
GO
PANTHER
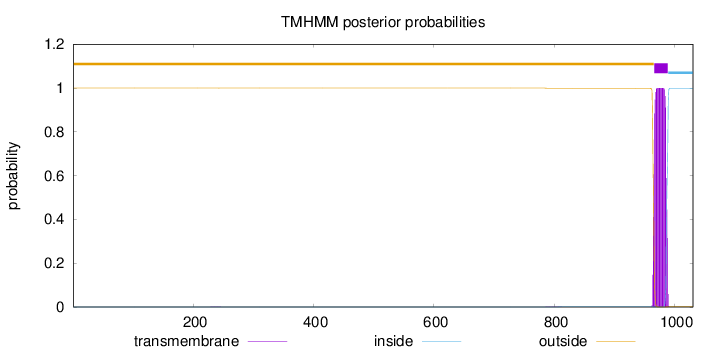
Topology
Length:
1031
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.28596
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.00019
outside
1 - 966
TMhelix
967 - 989
inside
990 - 1031
Population Genetic Test Statistics
Pi
278.695421
Theta
163.957856
Tajima's D
2.220787
CLR
0.367983
CSRT
0.911654417279136
Interpretation
Uncertain
