Gene
KWMTBOMO05373 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002547
Annotation
Neurexin-4_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.597 Nuclear Reliability : 1.597
Sequence
CDS
ATGGCCAACAAAGGCAATGTGCGATGGGCATTTAATACACGTGAGTGGCAGCCAGCGAAAGAAGAAATATTATTGGCTTCAAGTTATATACAAAATGAAGAAAAACAGAGGATATCCAGATTTGTCTTCCAGAACGATGCTAAATCGTCACTTATTGGAAGACTCATGCTAAGAAAGTATGTACAGACCTCAACATCGACTCCTTATGATGAGATAAAATTTGGTAGAGATGACCGCGGGAAACCGTTTTTGCTTAATCCTATTGATTTGAAGATAAGTTTCAATGTTTCTCACCAAGGAGACTTTGTGGTTCTAGCTGGATGCAAAGATTGGAATGTGGGAATTGACGTCATGAAAATTGAGCCACCTGTGAACAAGAATATCCCAGAGTTCTTTCGGATCATGGCCAGGCAGTTCTCACCACGAGAATGGTCGACTATACGCAGTTTTCCAACTGAGTTAGAGCAGATTGCTTGTTTTTACCGTATATGGTGCTTAAAAGAAAGTTACGTAAAGAATGTGGGGTTTGGTATAACTGTTCCTTTGGATGAAATCAGTTTTCAACTGAAGACTTTACATTTAGAAATGGACAAAATTGTTACTGATACTGTTCTCTATGAGAGGAATGTTTTGAAGGAGGATTGGATCTTTGAAGAGACATTACTCGATGAGAAGCATGCTGTAGCTGTGTCTTTGCAAAAGAAAGATAATTCAACAGATGCTCCAGTATGCTATAAGGTTTTGTCTTATCAAGAGCTGGTTGAGGAAGCCAAGTCTCTGTGTAGTACTGATGAAACATTTAGTACAAACGCTTGGACTGCTTCGGAGAGTGATTTCGATCAGCAACTGATTGTTGACTTTGGCAGTGTTAAGAACATAACCAGGATCGCAGTACAAGGTCGGGCTCATTCCCAAGAGTTTGTCCAGGAATACCATATCAGCTATGGATCGAATGGCCTAGATTACGTACAGTATAAAGCTCCTGGTGGTGAAGTGAAGATGTTTCACGGGAATCAAGACGGTGACACAGTGCGAAAAAACGAATTCGAAGTGCCTATAATAGCTCAATATTTGCGCATTAACCCGATGAGATGGCGGGACAAAATATCTATGAGAGTGGAAGTTTACGGTTGCGATTACGTCGCCGACACTCTGTACTTCAACGGGTCGTCGCTAATAAAAATGGACCTGCTCCGGGACCCGATATCGGCGTCCCGGGAAGTGATCCGGTTCCGGCTGAAGACGAGCACGGCGTCGGGCGCGCTGCTGTACTCGCGCGGCACGCAGGGCGACTACCTGGCGCTGCAGCTCAGGGACAACCGGCTCGTGCTCAACATTGATTTGGCGCCCAAGTGA
Protein
MANKGNVRWAFNTREWQPAKEEILLASSYIQNEEKQRISRFVFQNDAKSSLIGRLMLRKYVQTSTSTPYDEIKFGRDDRGKPFLLNPIDLKISFNVSHQGDFVVLAGCKDWNVGIDVMKIEPPVNKNIPEFFRIMARQFSPREWSTIRSFPTELEQIACFYRIWCLKESYVKNVGFGITVPLDEISFQLKTLHLEMDKIVTDTVLYERNVLKEDWIFEETLLDEKHAVAVSLQKKDNSTDAPVCYKVLSYQELVEEAKSLCSTDETFSTNAWTASESDFDQQLIVDFGSVKNITRIAVQGRAHSQEFVQEYHISYGSNGLDYVQYKAPGGEVKMFHGNQDGDTVRKNEFEVPIIAQYLRINPMRWRDKISMRVEVYGCDYVADTLYFNGSSLIKMDLLRDPISASREVIRFRLKTSTASGALLYSRGTQGDYLALQLRDNRLVLNIDLAPK
Summary
Uniprot
EMBL
BABH01011183
BABH01011184
BABH01011185
BABH01011186
ODYU01011424
SOQ57153.1
+ More
KQ458714 KPJ05430.1 JTDY01002332 KOB71646.1 AXCM01004161 KB631644 ERL84947.1 APCN01000208 QCYY01003086 ROT65426.1 LRGB01000781 KZS16007.1 CCAG010014702 JRES01001180 KNC24750.1 AXCN02000889 AXCN02000890 ATLV01024605 KE525352 KFB51771.1
KQ458714 KPJ05430.1 JTDY01002332 KOB71646.1 AXCM01004161 KB631644 ERL84947.1 APCN01000208 QCYY01003086 ROT65426.1 LRGB01000781 KZS16007.1 CCAG010014702 JRES01001180 KNC24750.1 AXCN02000889 AXCN02000890 ATLV01024605 KE525352 KFB51771.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
PDB
2C43
E-value=1.18682e-51,
Score=514
Ontologies
GO
PANTHER
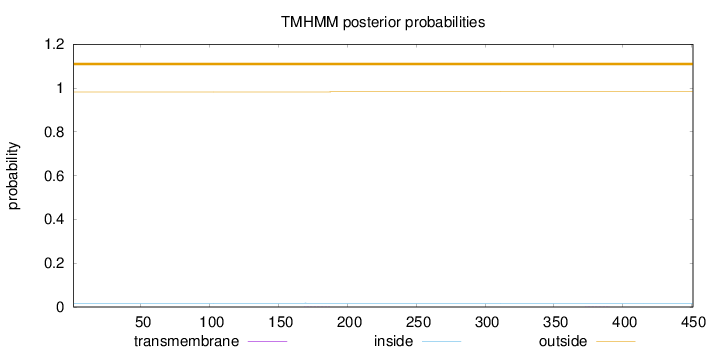
Topology
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00898999999999998
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.01765
outside
1 - 451
Population Genetic Test Statistics
Pi
243.080936
Theta
36.026817
Tajima's D
0.376756
CLR
1.76533
CSRT
0.481625918704065
Interpretation
Uncertain
