Gene
KWMTBOMO05371
Pre Gene Modal
BGIBMGA002546
Annotation
PREDICTED:_peptide_deformylase?_mitochondrial-like_isoform_X2_[Bombyx_mori]
Full name
Peptide deformylase
Location in the cell
Mitochondrial Reliability : 1.685
Sequence
CDS
ATGAAAATAACAAGGAAAATATTAAACTGGTATGCTAGATTAAGTCCTAAACACGGAGTATCATTACCTCCATATAATCATGTTGTTCAAGTTGGTGATCCAACATTACGTAAAGTTTCCGAGCCGGTACCAATAGAAAATATCAAAACCAAAGAAATACAGACGCTTATACTGAAATTACGTTTCGTTATGAACAAATACAAGTCAGTAGGTATGTCGGCTCCACAAATTGGTGTAAATATGAGAATATTTGTGATGCAACTGAATCCATTGCAACTTGCTAATGTACCCCAAGCAATTGTTAAATCAAGGGGCATGGAAGTGATTCCGTTTACTGTATTTGTCAATCCCACATTGAAAGTTTTGAACTATAACAAAGTGATATTGTCGGAGGGCTGTGAAAGTGTACAAGGATATACTGCTGATGTACCTAGATATAAAGAAATACAAATTTCAGGATACAATGAAGATGGAAATCCCACATCCAACACTTATAGAGACTGGGCTGCGAGAGTAGCTCAGCATGAGATTGAGCACTTAGATGGCAAACTCTATGTGGATATAATGGACCGAAAGACGATGTCCTGTGTATGCTGGGAGGAAGTTAATTTATCGAAAGGAAAACTTGCTATACCGTTTTCGCCTGAATAA
Protein
MKITRKILNWYARLSPKHGVSLPPYNHVVQVGDPTLRKVSEPVPIENIKTKEIQTLILKLRFVMNKYKSVGMSAPQIGVNMRIFVMQLNPLQLANVPQAIVKSRGMEVIPFTVFVNPTLKVLNYNKVILSEGCESVQGYTADVPRYKEIQISGYNEDGNPTSNTYRDWAARVAQHEIEHLDGKLYVDIMDRKTMSCVCWEEVNLSKGKLAIPFSPE
Summary
Description
Removes the formyl group from the N-terminal Met of newly synthesized proteins.
Catalytic Activity
H2O + N-terminal N-formyl-L-methionyl-[peptide] = formate + N-terminal L-methionyl-[peptide]
Similarity
Belongs to the polypeptide deformylase family.
Feature
chain Peptide deformylase
Uniprot
H9IZ61
A0A0L7K3Z8
A0A0L7LCS2
A0A2A4J7N4
A0A2H1V332
A0A194QIU2
+ More
A0A0N1PHI8 A0A3S2PCF2 A0A212EUW3 A0A2M4AZ14 W5J735 A0A0K8TSH0 A0A182FSV1 A0A182IRX0 B4KD11 A0A1W4WYC2 B3LWH4 A0A0M3QXU2 A0A182NFZ9 A0A182SKK3 A0A1Y1KDG8 A0A182MI78 A0A182UM64 A0A182WUZ8 A0A182I8M3 A0A182RRU1 A0A182LAI1 A0A1W4VZ62 A0A182P2P8 B4PKR6 A0A182W141 B4HIR6 A0A182TD21 A0A182Q0Z8 Q7QFS8 B4GLS6 Q9VGY2 A0A1Q3FAP0 Q293Q5 A0A3B0JTR2 B4LZJ6 A0A084W786 B3P1L6 A0A182XX55 D6X4P8 B4JUW6 B0WV60 B4NAZ0 B4QV62 A0A1Y1KGL9 A0A182G4M9 A0A1A9WDV1 A0A1S4EVE6 A0A023ELV7 A0A1I8PH39 Q17PR2 A0A1J1HVD8 A0A1B6G2U2 A0A1A9XWV4 A0A0K8V9R6 A0A1B6H4U6 A0A293LML5 A0A034WSX3 W4XDN7 A0A1B6HR30 A0A1I8NID8 A0A1B6E7B0 A0A0P4WMV8 A0A0A1WZD3 A0A1B0ACX5 E9H2A8 A0A069DQT1 A0A023F2A0 A0A1W4VYY0 A0A1B0FE35 A0A336MUU5 A0A195CRX4 A0A1B0B2E8 A0A164XAB7 A0A0J7K813 A0A1B6LR49 A0A336KR72 Q4V5F8 A0A2J7RJ01 T1IFZ8 E2AB28 A0A1S3DJF6 A0A1L8DRT9 Q8INL3 A0A195BPF3 A0A3Q2DH33 B4QV61 A0A0U2T6T6 B4GLS5 A0A158NZF2 Q293Q6 B4HIR5 A0A2G9QEX1 A0A151ITD0 A0A0P4VPU8
A0A0N1PHI8 A0A3S2PCF2 A0A212EUW3 A0A2M4AZ14 W5J735 A0A0K8TSH0 A0A182FSV1 A0A182IRX0 B4KD11 A0A1W4WYC2 B3LWH4 A0A0M3QXU2 A0A182NFZ9 A0A182SKK3 A0A1Y1KDG8 A0A182MI78 A0A182UM64 A0A182WUZ8 A0A182I8M3 A0A182RRU1 A0A182LAI1 A0A1W4VZ62 A0A182P2P8 B4PKR6 A0A182W141 B4HIR6 A0A182TD21 A0A182Q0Z8 Q7QFS8 B4GLS6 Q9VGY2 A0A1Q3FAP0 Q293Q5 A0A3B0JTR2 B4LZJ6 A0A084W786 B3P1L6 A0A182XX55 D6X4P8 B4JUW6 B0WV60 B4NAZ0 B4QV62 A0A1Y1KGL9 A0A182G4M9 A0A1A9WDV1 A0A1S4EVE6 A0A023ELV7 A0A1I8PH39 Q17PR2 A0A1J1HVD8 A0A1B6G2U2 A0A1A9XWV4 A0A0K8V9R6 A0A1B6H4U6 A0A293LML5 A0A034WSX3 W4XDN7 A0A1B6HR30 A0A1I8NID8 A0A1B6E7B0 A0A0P4WMV8 A0A0A1WZD3 A0A1B0ACX5 E9H2A8 A0A069DQT1 A0A023F2A0 A0A1W4VYY0 A0A1B0FE35 A0A336MUU5 A0A195CRX4 A0A1B0B2E8 A0A164XAB7 A0A0J7K813 A0A1B6LR49 A0A336KR72 Q4V5F8 A0A2J7RJ01 T1IFZ8 E2AB28 A0A1S3DJF6 A0A1L8DRT9 Q8INL3 A0A195BPF3 A0A3Q2DH33 B4QV61 A0A0U2T6T6 B4GLS5 A0A158NZF2 Q293Q6 B4HIR5 A0A2G9QEX1 A0A151ITD0 A0A0P4VPU8
EC Number
3.5.1.88
Pubmed
19121390
26227816
26354079
22118469
20920257
23761445
+ More
26369729 17994087 28004739 20966253 17550304 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18057021 24438588 25244985 18362917 19820115 26483478 17510324 24945155 25348373 25315136 25830018 21292972 26334808 25474469 20798317 26621068 21347285 27129103
26369729 17994087 28004739 20966253 17550304 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18057021 24438588 25244985 18362917 19820115 26483478 17510324 24945155 25348373 25315136 25830018 21292972 26334808 25474469 20798317 26621068 21347285 27129103
EMBL
BABH01011178
JTDY01011199
KOB57385.1
JTDY01001630
KOB73303.1
NWSH01002504
+ More
PCG68137.1 ODYU01000446 SOQ35231.1 KQ458714 KPJ05432.1 KQ460864 KPJ11809.1 RSAL01000103 RVE47418.1 AGBW02012297 OWR45247.1 GGFK01012696 MBW46017.1 ADMH02002125 ETN58675.1 GDAI01000520 JAI17083.1 CH933806 EDW13781.1 CH902617 EDV41568.1 CP012526 ALC46523.1 GEZM01087669 JAV58548.1 AXCM01001964 APCN01003089 CM000160 EDW96825.1 CH480815 EDW42713.1 AXCN02000343 AAAB01008844 EAA06035.4 CH479185 EDW38500.1 AE014297 BT015988 AAF54540.2 AAV36873.1 GFDL01010425 JAV24620.1 CM000070 EAL29159.1 OUUW01000008 SPP83792.1 CH940650 EDW67135.1 KRF83128.1 ATLV01021173 KE525314 KFB46080.1 CH954181 EDV49615.1 KQ971380 EEZ97665.1 CH916374 EDV91286.1 DS232119 EDS35399.1 CH964232 EDW80954.1 CM000364 EDX13468.1 GEZM01087672 JAV58546.1 JXUM01042548 KQ561329 KXJ78959.1 GAPW01003341 JAC10257.1 CH477190 EAT48689.1 CVRI01000019 CRK90454.1 GECZ01013009 JAS56760.1 GDHF01016688 JAI35626.1 GECZ01000076 JAS69693.1 GFWV01004361 MAA29091.1 GAKP01000296 JAC58656.1 AAGJ04150145 GECU01030631 JAS77075.1 GEDC01003503 JAS33795.1 GDRN01060426 JAI65367.1 GBXI01010442 JAD03850.1 GL732585 EFX74141.1 GBGD01002892 JAC85997.1 GBBI01003613 JAC15099.1 CCAG010018329 UFQT01002185 SSX32879.1 KQ977349 KYN03390.1 JXJN01007618 LRGB01001005 KZS14023.1 LBMM01012204 KMQ86351.1 GEBQ01013822 JAT26155.1 UFQS01000859 UFQT01000859 SSX07326.1 SSX27668.1 BT022698 AAY55114.1 NEVH01003009 PNF40818.1 ACPB03011443 GL438234 EFN69304.1 GFDF01004963 JAV09121.1 AAN13481.1 KQ976433 KYM87332.1 EDX13467.1 KT754454 ALS04288.1 EDW38499.1 ADTU01004571 EAL29158.1 EDW42712.1 KZ027616 PIO13613.1 KQ981018 KYN10259.1 GDKW01002086 JAI54509.1
PCG68137.1 ODYU01000446 SOQ35231.1 KQ458714 KPJ05432.1 KQ460864 KPJ11809.1 RSAL01000103 RVE47418.1 AGBW02012297 OWR45247.1 GGFK01012696 MBW46017.1 ADMH02002125 ETN58675.1 GDAI01000520 JAI17083.1 CH933806 EDW13781.1 CH902617 EDV41568.1 CP012526 ALC46523.1 GEZM01087669 JAV58548.1 AXCM01001964 APCN01003089 CM000160 EDW96825.1 CH480815 EDW42713.1 AXCN02000343 AAAB01008844 EAA06035.4 CH479185 EDW38500.1 AE014297 BT015988 AAF54540.2 AAV36873.1 GFDL01010425 JAV24620.1 CM000070 EAL29159.1 OUUW01000008 SPP83792.1 CH940650 EDW67135.1 KRF83128.1 ATLV01021173 KE525314 KFB46080.1 CH954181 EDV49615.1 KQ971380 EEZ97665.1 CH916374 EDV91286.1 DS232119 EDS35399.1 CH964232 EDW80954.1 CM000364 EDX13468.1 GEZM01087672 JAV58546.1 JXUM01042548 KQ561329 KXJ78959.1 GAPW01003341 JAC10257.1 CH477190 EAT48689.1 CVRI01000019 CRK90454.1 GECZ01013009 JAS56760.1 GDHF01016688 JAI35626.1 GECZ01000076 JAS69693.1 GFWV01004361 MAA29091.1 GAKP01000296 JAC58656.1 AAGJ04150145 GECU01030631 JAS77075.1 GEDC01003503 JAS33795.1 GDRN01060426 JAI65367.1 GBXI01010442 JAD03850.1 GL732585 EFX74141.1 GBGD01002892 JAC85997.1 GBBI01003613 JAC15099.1 CCAG010018329 UFQT01002185 SSX32879.1 KQ977349 KYN03390.1 JXJN01007618 LRGB01001005 KZS14023.1 LBMM01012204 KMQ86351.1 GEBQ01013822 JAT26155.1 UFQS01000859 UFQT01000859 SSX07326.1 SSX27668.1 BT022698 AAY55114.1 NEVH01003009 PNF40818.1 ACPB03011443 GL438234 EFN69304.1 GFDF01004963 JAV09121.1 AAN13481.1 KQ976433 KYM87332.1 EDX13467.1 KT754454 ALS04288.1 EDW38499.1 ADTU01004571 EAL29158.1 EDW42712.1 KZ027616 PIO13613.1 KQ981018 KYN10259.1 GDKW01002086 JAI54509.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000283053
+ More
UP000007151 UP000000673 UP000069272 UP000075880 UP000009192 UP000192223 UP000007801 UP000092553 UP000075884 UP000075901 UP000075883 UP000075903 UP000076407 UP000075840 UP000075900 UP000075882 UP000192221 UP000075885 UP000002282 UP000075920 UP000001292 UP000075902 UP000075886 UP000007062 UP000008744 UP000000803 UP000001819 UP000268350 UP000008792 UP000030765 UP000008711 UP000076408 UP000007266 UP000001070 UP000002320 UP000007798 UP000000304 UP000069940 UP000249989 UP000091820 UP000095300 UP000008820 UP000183832 UP000092443 UP000007110 UP000095301 UP000092445 UP000000305 UP000092444 UP000078542 UP000092460 UP000076858 UP000036403 UP000235965 UP000015103 UP000000311 UP000079169 UP000078540 UP000265020 UP000005205 UP000078492
UP000007151 UP000000673 UP000069272 UP000075880 UP000009192 UP000192223 UP000007801 UP000092553 UP000075884 UP000075901 UP000075883 UP000075903 UP000076407 UP000075840 UP000075900 UP000075882 UP000192221 UP000075885 UP000002282 UP000075920 UP000001292 UP000075902 UP000075886 UP000007062 UP000008744 UP000000803 UP000001819 UP000268350 UP000008792 UP000030765 UP000008711 UP000076408 UP000007266 UP000001070 UP000002320 UP000007798 UP000000304 UP000069940 UP000249989 UP000091820 UP000095300 UP000008820 UP000183832 UP000092443 UP000007110 UP000095301 UP000092445 UP000000305 UP000092444 UP000078542 UP000092460 UP000076858 UP000036403 UP000235965 UP000015103 UP000000311 UP000079169 UP000078540 UP000265020 UP000005205 UP000078492
Interpro
Gene 3D
ProteinModelPortal
H9IZ61
A0A0L7K3Z8
A0A0L7LCS2
A0A2A4J7N4
A0A2H1V332
A0A194QIU2
+ More
A0A0N1PHI8 A0A3S2PCF2 A0A212EUW3 A0A2M4AZ14 W5J735 A0A0K8TSH0 A0A182FSV1 A0A182IRX0 B4KD11 A0A1W4WYC2 B3LWH4 A0A0M3QXU2 A0A182NFZ9 A0A182SKK3 A0A1Y1KDG8 A0A182MI78 A0A182UM64 A0A182WUZ8 A0A182I8M3 A0A182RRU1 A0A182LAI1 A0A1W4VZ62 A0A182P2P8 B4PKR6 A0A182W141 B4HIR6 A0A182TD21 A0A182Q0Z8 Q7QFS8 B4GLS6 Q9VGY2 A0A1Q3FAP0 Q293Q5 A0A3B0JTR2 B4LZJ6 A0A084W786 B3P1L6 A0A182XX55 D6X4P8 B4JUW6 B0WV60 B4NAZ0 B4QV62 A0A1Y1KGL9 A0A182G4M9 A0A1A9WDV1 A0A1S4EVE6 A0A023ELV7 A0A1I8PH39 Q17PR2 A0A1J1HVD8 A0A1B6G2U2 A0A1A9XWV4 A0A0K8V9R6 A0A1B6H4U6 A0A293LML5 A0A034WSX3 W4XDN7 A0A1B6HR30 A0A1I8NID8 A0A1B6E7B0 A0A0P4WMV8 A0A0A1WZD3 A0A1B0ACX5 E9H2A8 A0A069DQT1 A0A023F2A0 A0A1W4VYY0 A0A1B0FE35 A0A336MUU5 A0A195CRX4 A0A1B0B2E8 A0A164XAB7 A0A0J7K813 A0A1B6LR49 A0A336KR72 Q4V5F8 A0A2J7RJ01 T1IFZ8 E2AB28 A0A1S3DJF6 A0A1L8DRT9 Q8INL3 A0A195BPF3 A0A3Q2DH33 B4QV61 A0A0U2T6T6 B4GLS5 A0A158NZF2 Q293Q6 B4HIR5 A0A2G9QEX1 A0A151ITD0 A0A0P4VPU8
A0A0N1PHI8 A0A3S2PCF2 A0A212EUW3 A0A2M4AZ14 W5J735 A0A0K8TSH0 A0A182FSV1 A0A182IRX0 B4KD11 A0A1W4WYC2 B3LWH4 A0A0M3QXU2 A0A182NFZ9 A0A182SKK3 A0A1Y1KDG8 A0A182MI78 A0A182UM64 A0A182WUZ8 A0A182I8M3 A0A182RRU1 A0A182LAI1 A0A1W4VZ62 A0A182P2P8 B4PKR6 A0A182W141 B4HIR6 A0A182TD21 A0A182Q0Z8 Q7QFS8 B4GLS6 Q9VGY2 A0A1Q3FAP0 Q293Q5 A0A3B0JTR2 B4LZJ6 A0A084W786 B3P1L6 A0A182XX55 D6X4P8 B4JUW6 B0WV60 B4NAZ0 B4QV62 A0A1Y1KGL9 A0A182G4M9 A0A1A9WDV1 A0A1S4EVE6 A0A023ELV7 A0A1I8PH39 Q17PR2 A0A1J1HVD8 A0A1B6G2U2 A0A1A9XWV4 A0A0K8V9R6 A0A1B6H4U6 A0A293LML5 A0A034WSX3 W4XDN7 A0A1B6HR30 A0A1I8NID8 A0A1B6E7B0 A0A0P4WMV8 A0A0A1WZD3 A0A1B0ACX5 E9H2A8 A0A069DQT1 A0A023F2A0 A0A1W4VYY0 A0A1B0FE35 A0A336MUU5 A0A195CRX4 A0A1B0B2E8 A0A164XAB7 A0A0J7K813 A0A1B6LR49 A0A336KR72 Q4V5F8 A0A2J7RJ01 T1IFZ8 E2AB28 A0A1S3DJF6 A0A1L8DRT9 Q8INL3 A0A195BPF3 A0A3Q2DH33 B4QV61 A0A0U2T6T6 B4GLS5 A0A158NZF2 Q293Q6 B4HIR5 A0A2G9QEX1 A0A151ITD0 A0A0P4VPU8
PDB
3G5P
E-value=2.04962e-36,
Score=379
Ontologies
GO
PANTHER
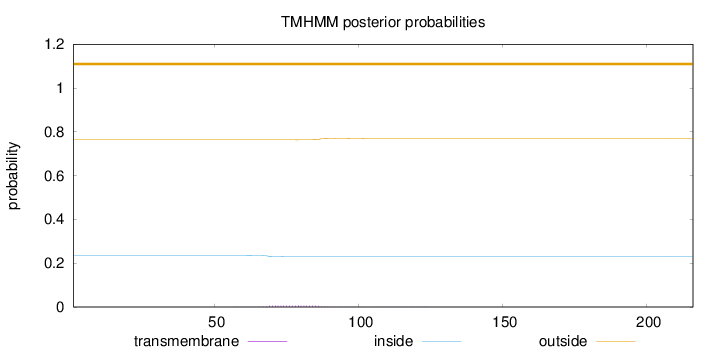
Topology
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14849
Exp number, first 60 AAs:
0.00244
Total prob of N-in:
0.23655
outside
1 - 216
Population Genetic Test Statistics
Pi
177.47847
Theta
159.837141
Tajima's D
0.347067
CLR
0
CSRT
0.473226338683066
Interpretation
Uncertain
