Gene
KWMTBOMO05370 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002376
Annotation
PREDICTED:_3-hydroxyisobutyryl-CoA_hydrolase?_mitochondrial_[Papilio_xuthus]
Full name
3-hydroxyisobutyryl-CoA hydrolase, mitochondrial
Alternative Name
3-hydroxyisobutyryl-coenzyme A hydrolase
Location in the cell
Mitochondrial Reliability : 2.332
Sequence
CDS
ATGTATAGTTTAATAAAACTTAAACAAGCAACATTGCCTTTGTTAAGAAGGACAATGTCTTCCCAAGAACCTGATGTTTTATTTGAAGCATTAAACAATGCTGGCATAGTAACCCTCAATCGACCCAAAGCATTAAATTCCCTCAACACATCTATGGTGTCAAAATTATTACCACAACTTCAAGAATGGGAGAAATCAAAGTCTCTGGTTATAATTAAGGGTGCTGGAGAGAAAGCTTTCTGTGCTGGTGGTGATGTGAAAGCTGCCATAGACAAAATGGAAGGGCCTAGGTTCTTCCATACGGAATACAATGTTAATTACTTGATTGGAAACTATAAAATCCCATATATTGCATTTATAAATGGAATCACAATGGGAGGAGGTCTTGGACTCTCAGTCCATGGCAGATACAGAGTAGCAACAGAGAAGACATTAATAGCCATGCCAGAGACTAAGATAGGACTATTCCCAGATGTTGGTGGATCATTCTTTTTACCTCGTTTACAAGTTAATTTGGGATTGTACTTAGGGTTGACCGGTGACAGATTGAAAGGTAAAGACGTTGTGAAAGCTGGAATAGCTACACACTTTGTACCAAGCAAACGTCTGTATGAACTCGAAATGCTCCTTTCTCGGTGCGCTAATGATATAGAAGTTAATTCACTACTCAATAAATTCCACGAACCCTCGGAGGACTTCTCACTTGCATCAAATATCAAGCATATAAACTACTGTTTTGCGGCTTCGACTGTTGAAGAGATCATTGAGCGACTGGAGAAAGTTCAAAATGAATGGTCCGTTAAGACTTTAGAGACTCTTCATCAAATGTGTCCTGGCTCTCTCAAGATTACACTGCGAGCCCTCCAACGAGGCGCTCAGCTGGACCTGGGCCAGTGTCTACAGATGGAGTACAGACTGGCCTGCCGAGCCACTGAGAACCATGATTTCCCTGAAGGTGTGAGAGCTCTGCTCATCGACAAGGACAACAAGCCCCAGTGGAACCCCAAGTCTCTGGCCGAAGCTGATGATGAATATGTCGAGAGTTATTTCAAGAGGCTGCCCGAAGACAAAGAACTCAAATTCTTCGACTCCAAACTCTAA
Protein
MYSLIKLKQATLPLLRRTMSSQEPDVLFEALNNAGIVTLNRPKALNSLNTSMVSKLLPQLQEWEKSKSLVIIKGAGEKAFCAGGDVKAAIDKMEGPRFFHTEYNVNYLIGNYKIPYIAFINGITMGGGLGLSVHGRYRVATEKTLIAMPETKIGLFPDVGGSFFLPRLQVNLGLYLGLTGDRLKGKDVVKAGIATHFVPSKRLYELEMLLSRCANDIEVNSLLNKFHEPSEDFSLASNIKHINYCFAASTVEEIIERLEKVQNEWSVKTLETLHQMCPGSLKITLRALQRGAQLDLGQCLQMEYRLACRATENHDFPEGVRALLIDKDNKPQWNPKSLAEADDEYVESYFKRLPEDKELKFFDSKL
Summary
Description
Hydrolyzes 3-hydroxyisobutyryl-CoA (HIBYL-CoA), a saline catabolite.
Catalytic Activity
3-hydroxy-2-methylpropanoyl-CoA + H2O = 3-hydroxy-2-methylpropanoate + CoA + H(+)
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
H9IYP2
A0A2H1V310
A0A3S2NCD3
A0A2A4J756
A0A0N1ID25
A0A194QIQ0
+ More
A0A068FL83 A0A0L7KSG9 A0A0L7K3Z7 V5I815 A0A1I8PG25 A0A1I8PGA5 A0A1I8PG41 A0A1A9VLV9 A0A1I8MUP3 A0A1I8MUP0 A0A0L0BZ30 A0A0L7R4N1 T1PCM3 T1PC23 D3TLD0 D6X3R7 A0A1A9W2U7 Q960K8 Q86BP1 A0A0R1E9B9 B4PS39 A0A1Y1KBE0 B3MSU0 A0A0P8XE63 B3P0V9 A0A195CSV4 A0A1W4VF24 A0A1W4VS18 B4HM22 E2A6R8 A0A2M3Z9X9 A0A2P8YBE1 B4QYI3 A0A2M3ZAC7 A0A158NIN6 A0A2M3Z9W8 A0A154P6H7 U4U241 A0A2M4AKP7 A0A151JQI3 A0A0Q9X4J3 B4KAQ0 B4NAU1 W5JCR8 A0A2M4BRS1 B4M5Z4 A0A0M4EGI5 Q7PH36 E9IZM3 A0A1I8JW73 A0A1A9YFT1 A0A182JSH7 A0A0Q9WBF2 A0A3L8DI43 W8BDX4 A0A182L7K7 A0A182V5D2 A0A026WMN3 A0A336LWI3 U5EWB6 A0A3B0JJ12 E2BCN8 A0A0K8V6Q1 Q29BE1 B4GNT9 A0A0R3NKE4 A0A0K8UEN6 F4WDJ8 A0A0K8VM74 A0A182F774 A0A023EQY7 A0A182PNM7 A0A1S4FP05 Q16U65 A0A182Y753 A0A151WG81 A0A1Y9HAB9 A0A0P4WCR1 A0A182RID0 B4JU41 A0A182T1N3 A0A034VRP5 A0A034VQY3 A0A182HCD9 A0A084VCI1 A0A088AET4 A0A3R7QX48 A0A0T6B2K3 A0A182TUV5 K7IWX1 A0A182N5K7 A0A182I7Z8 A0A1B6CRN6
A0A068FL83 A0A0L7KSG9 A0A0L7K3Z7 V5I815 A0A1I8PG25 A0A1I8PGA5 A0A1I8PG41 A0A1A9VLV9 A0A1I8MUP3 A0A1I8MUP0 A0A0L0BZ30 A0A0L7R4N1 T1PCM3 T1PC23 D3TLD0 D6X3R7 A0A1A9W2U7 Q960K8 Q86BP1 A0A0R1E9B9 B4PS39 A0A1Y1KBE0 B3MSU0 A0A0P8XE63 B3P0V9 A0A195CSV4 A0A1W4VF24 A0A1W4VS18 B4HM22 E2A6R8 A0A2M3Z9X9 A0A2P8YBE1 B4QYI3 A0A2M3ZAC7 A0A158NIN6 A0A2M3Z9W8 A0A154P6H7 U4U241 A0A2M4AKP7 A0A151JQI3 A0A0Q9X4J3 B4KAQ0 B4NAU1 W5JCR8 A0A2M4BRS1 B4M5Z4 A0A0M4EGI5 Q7PH36 E9IZM3 A0A1I8JW73 A0A1A9YFT1 A0A182JSH7 A0A0Q9WBF2 A0A3L8DI43 W8BDX4 A0A182L7K7 A0A182V5D2 A0A026WMN3 A0A336LWI3 U5EWB6 A0A3B0JJ12 E2BCN8 A0A0K8V6Q1 Q29BE1 B4GNT9 A0A0R3NKE4 A0A0K8UEN6 F4WDJ8 A0A0K8VM74 A0A182F774 A0A023EQY7 A0A182PNM7 A0A1S4FP05 Q16U65 A0A182Y753 A0A151WG81 A0A1Y9HAB9 A0A0P4WCR1 A0A182RID0 B4JU41 A0A182T1N3 A0A034VRP5 A0A034VQY3 A0A182HCD9 A0A084VCI1 A0A088AET4 A0A3R7QX48 A0A0T6B2K3 A0A182TUV5 K7IWX1 A0A182N5K7 A0A182I7Z8 A0A1B6CRN6
EC Number
3.1.2.4
Pubmed
19121390
26354079
26385554
26227816
25315136
26108605
+ More
20353571 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 28004739 20798317 29403074 21347285 23537049 20920257 23761445 12364791 14747013 17210077 21282665 30249741 24495485 20966253 24508170 15632085 21719571 24945155 17510324 25244985 25348373 26483478 24438588 20075255
20353571 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 28004739 20798317 29403074 21347285 23537049 20920257 23761445 12364791 14747013 17210077 21282665 30249741 24495485 20966253 24508170 15632085 21719571 24945155 17510324 25244985 25348373 26483478 24438588 20075255
EMBL
BABH01011177
ODYU01000446
SOQ35230.1
RSAL01000103
RVE47419.1
NWSH01002810
+ More
PCG67498.1 KQ460864 KPJ11808.1 KQ458714 KPJ05433.1 KJ622098 AID66688.1 JTDY01006213 KOB66192.1 JTDY01011199 KOB57386.1 GALX01005475 JAB62991.1 JRES01001133 KNC25261.1 KQ414657 KOC65721.1 KA646486 AFP61115.1 KA646239 AFP60868.1 CCAG010005443 CCAG010005444 EZ422232 ADD18508.1 KQ971377 EEZ97382.1 AE014297 AY052009 AAF55181.2 AAK93433.1 BT126275 AAN13658.2 AEB39636.1 CM000160 KRK03767.1 EDW97459.2 GEZM01087220 JAV58832.1 CH902623 EDV30330.2 KPU72800.1 CH954181 EDV49007.1 KQ977306 KYN03607.1 CH480815 EDW42059.1 GL437197 EFN70862.1 GGFM01004596 MBW25347.1 PYGN01000732 PSN41571.1 CM000364 EDX12828.1 GGFM01004647 MBW25398.1 ADTU01017028 GGFM01004572 MBW25323.1 KQ434810 KZC06740.1 KB631615 ERL84686.1 GGFK01008044 MBW41365.1 KQ978673 KYN29336.1 CH933806 KRG02297.1 EDW16787.2 CH964232 EDW80905.2 ADMH02001534 ETN62147.1 GGFJ01006618 MBW55759.1 CH940652 EDW59070.2 CP012526 ALC47491.1 AAAB01008880 EAA44701.5 GL767232 EFZ13911.1 KRF78660.1 QOIP01000007 RLU20110.1 GAMC01018586 JAB87969.1 KK107152 EZA57173.1 UFQS01000235 UFQT01000235 SSX01793.1 SSX22170.1 GANO01003023 JAB56848.1 OUUW01000005 SPP80743.1 GL447336 EFN86548.1 GDHF01028827 GDHF01017702 JAI23487.1 JAI34612.1 CM000070 EAL27057.1 CH479186 EDW38822.1 KRS99748.1 GDHF01027554 GDHF01027239 JAI24760.1 JAI25075.1 GL888087 EGI67778.1 GDHF01012327 JAI39987.1 GAPW01002060 JAC11538.1 CH477631 EAT38068.1 KQ983189 KYQ46817.1 AXCN02001161 GDRN01080807 JAI62131.1 CH916374 EDV91011.1 GAKP01014766 GAKP01014764 JAC44188.1 GAKP01014762 JAC44190.1 JXUM01127484 KQ567197 KXJ69562.1 ATLV01010759 KE524612 KFB35675.1 QCYY01000974 ROT81432.1 LJIG01016113 KRT81576.1 AAZX01008308 APCN01002591 GEDC01021237 JAS16061.1
PCG67498.1 KQ460864 KPJ11808.1 KQ458714 KPJ05433.1 KJ622098 AID66688.1 JTDY01006213 KOB66192.1 JTDY01011199 KOB57386.1 GALX01005475 JAB62991.1 JRES01001133 KNC25261.1 KQ414657 KOC65721.1 KA646486 AFP61115.1 KA646239 AFP60868.1 CCAG010005443 CCAG010005444 EZ422232 ADD18508.1 KQ971377 EEZ97382.1 AE014297 AY052009 AAF55181.2 AAK93433.1 BT126275 AAN13658.2 AEB39636.1 CM000160 KRK03767.1 EDW97459.2 GEZM01087220 JAV58832.1 CH902623 EDV30330.2 KPU72800.1 CH954181 EDV49007.1 KQ977306 KYN03607.1 CH480815 EDW42059.1 GL437197 EFN70862.1 GGFM01004596 MBW25347.1 PYGN01000732 PSN41571.1 CM000364 EDX12828.1 GGFM01004647 MBW25398.1 ADTU01017028 GGFM01004572 MBW25323.1 KQ434810 KZC06740.1 KB631615 ERL84686.1 GGFK01008044 MBW41365.1 KQ978673 KYN29336.1 CH933806 KRG02297.1 EDW16787.2 CH964232 EDW80905.2 ADMH02001534 ETN62147.1 GGFJ01006618 MBW55759.1 CH940652 EDW59070.2 CP012526 ALC47491.1 AAAB01008880 EAA44701.5 GL767232 EFZ13911.1 KRF78660.1 QOIP01000007 RLU20110.1 GAMC01018586 JAB87969.1 KK107152 EZA57173.1 UFQS01000235 UFQT01000235 SSX01793.1 SSX22170.1 GANO01003023 JAB56848.1 OUUW01000005 SPP80743.1 GL447336 EFN86548.1 GDHF01028827 GDHF01017702 JAI23487.1 JAI34612.1 CM000070 EAL27057.1 CH479186 EDW38822.1 KRS99748.1 GDHF01027554 GDHF01027239 JAI24760.1 JAI25075.1 GL888087 EGI67778.1 GDHF01012327 JAI39987.1 GAPW01002060 JAC11538.1 CH477631 EAT38068.1 KQ983189 KYQ46817.1 AXCN02001161 GDRN01080807 JAI62131.1 CH916374 EDV91011.1 GAKP01014766 GAKP01014764 JAC44188.1 GAKP01014762 JAC44190.1 JXUM01127484 KQ567197 KXJ69562.1 ATLV01010759 KE524612 KFB35675.1 QCYY01000974 ROT81432.1 LJIG01016113 KRT81576.1 AAZX01008308 APCN01002591 GEDC01021237 JAS16061.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000037510
+ More
UP000095300 UP000078200 UP000095301 UP000037069 UP000053825 UP000092444 UP000007266 UP000091820 UP000000803 UP000002282 UP000007801 UP000008711 UP000078542 UP000192221 UP000001292 UP000000311 UP000245037 UP000000304 UP000005205 UP000076502 UP000030742 UP000078492 UP000009192 UP000007798 UP000000673 UP000008792 UP000092553 UP000007062 UP000076407 UP000092443 UP000075881 UP000279307 UP000075882 UP000075903 UP000053097 UP000268350 UP000008237 UP000001819 UP000008744 UP000007755 UP000069272 UP000075885 UP000008820 UP000076408 UP000075809 UP000075886 UP000075900 UP000001070 UP000075901 UP000069940 UP000249989 UP000030765 UP000005203 UP000283509 UP000075902 UP000002358 UP000075884 UP000075840
UP000095300 UP000078200 UP000095301 UP000037069 UP000053825 UP000092444 UP000007266 UP000091820 UP000000803 UP000002282 UP000007801 UP000008711 UP000078542 UP000192221 UP000001292 UP000000311 UP000245037 UP000000304 UP000005205 UP000076502 UP000030742 UP000078492 UP000009192 UP000007798 UP000000673 UP000008792 UP000092553 UP000007062 UP000076407 UP000092443 UP000075881 UP000279307 UP000075882 UP000075903 UP000053097 UP000268350 UP000008237 UP000001819 UP000008744 UP000007755 UP000069272 UP000075885 UP000008820 UP000076408 UP000075809 UP000075886 UP000075900 UP000001070 UP000075901 UP000069940 UP000249989 UP000030765 UP000005203 UP000283509 UP000075902 UP000002358 UP000075884 UP000075840
Interpro
SUPFAM
SSF52096
SSF52096
Gene 3D
ProteinModelPortal
H9IYP2
A0A2H1V310
A0A3S2NCD3
A0A2A4J756
A0A0N1ID25
A0A194QIQ0
+ More
A0A068FL83 A0A0L7KSG9 A0A0L7K3Z7 V5I815 A0A1I8PG25 A0A1I8PGA5 A0A1I8PG41 A0A1A9VLV9 A0A1I8MUP3 A0A1I8MUP0 A0A0L0BZ30 A0A0L7R4N1 T1PCM3 T1PC23 D3TLD0 D6X3R7 A0A1A9W2U7 Q960K8 Q86BP1 A0A0R1E9B9 B4PS39 A0A1Y1KBE0 B3MSU0 A0A0P8XE63 B3P0V9 A0A195CSV4 A0A1W4VF24 A0A1W4VS18 B4HM22 E2A6R8 A0A2M3Z9X9 A0A2P8YBE1 B4QYI3 A0A2M3ZAC7 A0A158NIN6 A0A2M3Z9W8 A0A154P6H7 U4U241 A0A2M4AKP7 A0A151JQI3 A0A0Q9X4J3 B4KAQ0 B4NAU1 W5JCR8 A0A2M4BRS1 B4M5Z4 A0A0M4EGI5 Q7PH36 E9IZM3 A0A1I8JW73 A0A1A9YFT1 A0A182JSH7 A0A0Q9WBF2 A0A3L8DI43 W8BDX4 A0A182L7K7 A0A182V5D2 A0A026WMN3 A0A336LWI3 U5EWB6 A0A3B0JJ12 E2BCN8 A0A0K8V6Q1 Q29BE1 B4GNT9 A0A0R3NKE4 A0A0K8UEN6 F4WDJ8 A0A0K8VM74 A0A182F774 A0A023EQY7 A0A182PNM7 A0A1S4FP05 Q16U65 A0A182Y753 A0A151WG81 A0A1Y9HAB9 A0A0P4WCR1 A0A182RID0 B4JU41 A0A182T1N3 A0A034VRP5 A0A034VQY3 A0A182HCD9 A0A084VCI1 A0A088AET4 A0A3R7QX48 A0A0T6B2K3 A0A182TUV5 K7IWX1 A0A182N5K7 A0A182I7Z8 A0A1B6CRN6
A0A068FL83 A0A0L7KSG9 A0A0L7K3Z7 V5I815 A0A1I8PG25 A0A1I8PGA5 A0A1I8PG41 A0A1A9VLV9 A0A1I8MUP3 A0A1I8MUP0 A0A0L0BZ30 A0A0L7R4N1 T1PCM3 T1PC23 D3TLD0 D6X3R7 A0A1A9W2U7 Q960K8 Q86BP1 A0A0R1E9B9 B4PS39 A0A1Y1KBE0 B3MSU0 A0A0P8XE63 B3P0V9 A0A195CSV4 A0A1W4VF24 A0A1W4VS18 B4HM22 E2A6R8 A0A2M3Z9X9 A0A2P8YBE1 B4QYI3 A0A2M3ZAC7 A0A158NIN6 A0A2M3Z9W8 A0A154P6H7 U4U241 A0A2M4AKP7 A0A151JQI3 A0A0Q9X4J3 B4KAQ0 B4NAU1 W5JCR8 A0A2M4BRS1 B4M5Z4 A0A0M4EGI5 Q7PH36 E9IZM3 A0A1I8JW73 A0A1A9YFT1 A0A182JSH7 A0A0Q9WBF2 A0A3L8DI43 W8BDX4 A0A182L7K7 A0A182V5D2 A0A026WMN3 A0A336LWI3 U5EWB6 A0A3B0JJ12 E2BCN8 A0A0K8V6Q1 Q29BE1 B4GNT9 A0A0R3NKE4 A0A0K8UEN6 F4WDJ8 A0A0K8VM74 A0A182F774 A0A023EQY7 A0A182PNM7 A0A1S4FP05 Q16U65 A0A182Y753 A0A151WG81 A0A1Y9HAB9 A0A0P4WCR1 A0A182RID0 B4JU41 A0A182T1N3 A0A034VRP5 A0A034VQY3 A0A182HCD9 A0A084VCI1 A0A088AET4 A0A3R7QX48 A0A0T6B2K3 A0A182TUV5 K7IWX1 A0A182N5K7 A0A182I7Z8 A0A1B6CRN6
PDB
3BPT
E-value=7.67335e-90,
Score=842
Ontologies
PATHWAY
00280
Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
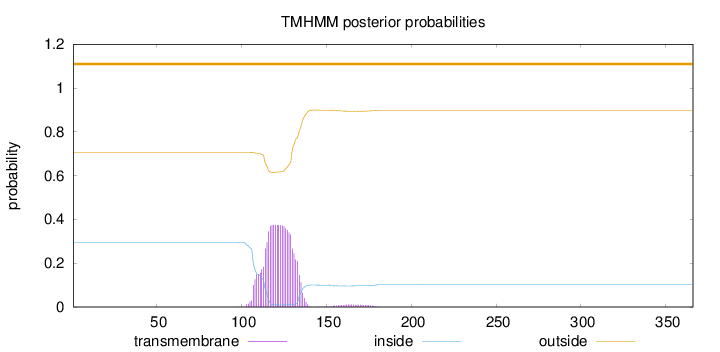
Topology
Subcellular location
Mitochondrion
Length:
366
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.33759
Exp number, first 60 AAs:
0.0013
Total prob of N-in:
0.29458
outside
1 - 366
Population Genetic Test Statistics
Pi
216.257796
Theta
178.029309
Tajima's D
0.537284
CLR
0.527515
CSRT
0.536173191340433
Interpretation
Uncertain
