Gene
KWMTBOMO05368
Pre Gene Modal
BGIBMGA002545
Annotation
_odorant_receptor_19_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
PlasmaMembrane Reliability : 4.931
Sequence
CDS
ATGGTTAACAAGATAAAGTCCTACTTAAGCACTTATGTGTTTGCGTCTATTACAATTCTAGTCAGTAATGGTATTATTGCATTTTTTCAAACAATTAACTCAGACGAACCATTCTTAGGAATAATAACAGCATGGCCAGATAAAACTGATACCTCAAAAACTGCTAGTTACGCAAGGATCGGTTTTTACTTATTCTGGTGCATTCACTTCTTTAGAATCTCAACAGTATTTGCTGTCATAGTATGTATCTTGATAAGTATCAAGTATCAGTATAAATTTTTATGCAGCTACTTTGAAAGTCTGAACAAAATCTTTGATGACGAAACTTCAAGCCATGAAGTTAAAGAGGCTGAATTTGAAAATGCTTTCTGTAATGGAATAAAAATACATACGCAAATAATTTGGTGTGTCAGAAGATGTCAAATTATGTGTCGGACCGTGTTCAGTGCTAATATTATGCTGGACACCTTTGTTCTCGTCATCTTGATGTTGGCGATGGTGAATTCAGAAAATGACTTCTACGGCTTGTGCTCACAAATGTCATCGGTACTGGTAACCGTTGTCCTGATGGCGTTCTTTATGTGGACAGCCGGAGATATTAACGTGCAGGCATCACAACTACCCGACGCCATCTACGGTTCGGGCTGGTACAACTGTCGTGGAAAGTCCTCAGCTCGTATTAGAAGCTTGGTCACTATCTCCATGAACAAAGCACAGGTGATTTTGACTCGTATGTATGGTAGTAACAAACAGTCTAACTACAATATGTTTAGCGGCGTATTCAGGGTGCAAATGGAAACTTGA
Protein
MVNKIKSYLSTYVFASITILVSNGIIAFFQTINSDEPFLGIITAWPDKTDTSKTASYARIGFYLFWCIHFFRISTVFAVIVCILISIKYQYKFLCSYFESLNKIFDDETSSHEVKEAEFENAFCNGIKIHTQIIWCVRRCQIMCRTVFSANIMLDTFVLVILMLAMVNSENDFYGLCSQMSSVLVTVVLMAFFMWTAGDINVQASQLPDAIYGSGWYNCRGKSSARIRSLVTISMNKAQVILTRMYGSNKQSNYNMFSGVFRVQMET
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Feature
chain Odorant receptor
Uniprot
A7E3G2
Q0EEG1
A0A076E7L0
A0A2K8GKX5
A0A076E611
E1ACG2
+ More
A0A0P1J400 A0A2K8GL46 A0A1B3B743 A0A2L1TG43 A0A2A4J5Z4 A0A2H1V355 A0A345BEX6 M1QVM8 A0A1V1WBU5 A0A223HD69 Q6A1J3 A0A0B5A1G3 A0A0B5CVM5 A0A2A4J6Z3 A0A097ITW0 A0A075T650 A0A097IYK2 A0A223HD63 H9A5Q2 A0A1B3B785 A0A223HDE8 A0A0K8TU59 A0A223HDG0 A0A2A4JBI2 A0A2L1TG66 A0A2H1V308 A0A1V1WC43 A0A0K8TVB3 A0A223HD37 A0A1B3B736 A0A345BEX5 A0A0S1TPQ8 A0A0B5GIB9 A0A097ITX4 A0A0B5CN09 A0A0V0J201 A0A345BEY1 U5NFU1 A0A0S1TPH0 A0A0S1TPK3 A0A075TD77 A0A0B5GUX0 A0A1W5T543 A0A2A4JBH1 A0A2A4JAB0 U5NJ19 A0A0E4B3W6 A0A223HD26 A0A223HD79 A0A223HDF4 C4B7V1 A0A2H1WY92 A0A146JW15 A0A1X9PBW3 A0A075T629 A0A0P1IVT1 A0A1B3B764 A0A2L1TG92 A0A223HD28 A0A2A4J9Y6 A0A075TD59 A0A075T6W5 A0A076E9B5 A0A3Q8HD97 A0A2K8GKR3 A0A1V1WBU9 A0A0P1IW73 A0A076E7I6 A0A0P1IVQ9 A7E3G3 A0A2H1WIS2 A0A2A4J6F5 A0A1B3P5N2 A0A146JYX6 A0A1Y9TJJ8 C4B7U9 A0A0N1PGA1 A0A1B3P5L3 A0A212FF61
A0A0P1J400 A0A2K8GL46 A0A1B3B743 A0A2L1TG43 A0A2A4J5Z4 A0A2H1V355 A0A345BEX6 M1QVM8 A0A1V1WBU5 A0A223HD69 Q6A1J3 A0A0B5A1G3 A0A0B5CVM5 A0A2A4J6Z3 A0A097ITW0 A0A075T650 A0A097IYK2 A0A223HD63 H9A5Q2 A0A1B3B785 A0A223HDE8 A0A0K8TU59 A0A223HDG0 A0A2A4JBI2 A0A2L1TG66 A0A2H1V308 A0A1V1WC43 A0A0K8TVB3 A0A223HD37 A0A1B3B736 A0A345BEX5 A0A0S1TPQ8 A0A0B5GIB9 A0A097ITX4 A0A0B5CN09 A0A0V0J201 A0A345BEY1 U5NFU1 A0A0S1TPH0 A0A0S1TPK3 A0A075TD77 A0A0B5GUX0 A0A1W5T543 A0A2A4JBH1 A0A2A4JAB0 U5NJ19 A0A0E4B3W6 A0A223HD26 A0A223HD79 A0A223HDF4 C4B7V1 A0A2H1WY92 A0A146JW15 A0A1X9PBW3 A0A075T629 A0A0P1IVT1 A0A1B3B764 A0A2L1TG92 A0A223HD28 A0A2A4J9Y6 A0A075TD59 A0A075T6W5 A0A076E9B5 A0A3Q8HD97 A0A2K8GKR3 A0A1V1WBU9 A0A0P1IW73 A0A076E7I6 A0A0P1IVQ9 A7E3G3 A0A2H1WIS2 A0A2A4J6F5 A0A1B3P5N2 A0A146JYX6 A0A1Y9TJJ8 C4B7U9 A0A0N1PGA1 A0A1B3P5L3 A0A212FF61
Pubmed
EMBL
BK005927
DAA05977.1
AB234349
BAF31189.1
KF487679
AII01077.1
+ More
KX585378 ARO70271.1 KF487693 AII01091.1 HM595406 LN885100 ADM32898.1 CUQ99389.1 LN885101 CUQ99390.1 KY225510 ARO70522.1 KT588127 AOE48037.1 KY707202 AVF19644.1 NWSH01002810 PCG67495.1 ODYU01000446 SOQ35229.1 MG816615 AXF48800.1 JX999589 AGG08879.1 GENK01000078 JAV45835.1 KY283671 AST36330.1 AJ748336 CAG38122.1 KJ542673 AJD81557.1 KM678325 AJE25898.1 PCG67496.1 KM597225 AIT69902.1 KF768701 AIG51895.1 KM655563 AIT72012.1 KY283667 AST36326.1 JN836702 AFC91742.1 KT588144 AOE48054.1 KY283743 AST36402.1 GCVX01000213 JAI18017.1 KY283746 AST36405.1 NWSH01002259 PCG68782.1 KY707225 AVF19667.1 SOQ35228.1 GENK01000077 JAV45836.1 GCVX01000220 JAI18010.1 KY283622 AST36281.1 KT588118 AOE48028.1 MG816614 AXF48799.1 KR935716 ALM26208.1 KM892406 AJF23821.1 KM597231 AIT69908.1 KM678332 AJE25905.1 GDKB01000055 JAP38441.1 MG816620 AXF48805.1 KC960481 AGY14592.1 KR935725 ALM26217.1 KR935757 ALM26249.1 KF768702 AIG51896.1 KM892399 AJF23814.1 KY825770 ARF06964.1 PCG68780.1 PCG68779.1 KC960482 AGY14593.1 LC002732 BAR43480.1 KY283619 AST36278.1 KY283678 AST36337.1 KY283757 AST36416.1 AB472100 BAH66317.1 ODYU01011979 SOQ58031.1 GEDO01000059 JAP88567.1 KX084488 ARO76443.1 KF768676 AIG51870.1 LN885119 CUQ99408.1 KT588150 AOE48060.1 KY707218 AVF19660.1 KY283628 AST36287.1 PCG68781.1 KF768677 AIG51871.1 KF768675 AIG51869.1 KF487691 AII01089.1 MF625594 AXY83421.1 KX585327 ARO70220.1 GENK01000081 JAV45832.1 LN885130 CUQ99419.1 KF487654 AII01052.1 LN885104 CUQ99393.1 BK005928 DAA05978.1 ODYU01008612 SOQ52344.1 NWSH01002695 PCG67665.1 KX655966 AOG12915.1 GEDO01000025 JAP88601.1 KX084495 ARO76450.1 AB472098 BAH66315.1 KQ460622 KPJ13512.1 KX655957 AOG12906.1 AGBW02008859 OWR52357.1
KX585378 ARO70271.1 KF487693 AII01091.1 HM595406 LN885100 ADM32898.1 CUQ99389.1 LN885101 CUQ99390.1 KY225510 ARO70522.1 KT588127 AOE48037.1 KY707202 AVF19644.1 NWSH01002810 PCG67495.1 ODYU01000446 SOQ35229.1 MG816615 AXF48800.1 JX999589 AGG08879.1 GENK01000078 JAV45835.1 KY283671 AST36330.1 AJ748336 CAG38122.1 KJ542673 AJD81557.1 KM678325 AJE25898.1 PCG67496.1 KM597225 AIT69902.1 KF768701 AIG51895.1 KM655563 AIT72012.1 KY283667 AST36326.1 JN836702 AFC91742.1 KT588144 AOE48054.1 KY283743 AST36402.1 GCVX01000213 JAI18017.1 KY283746 AST36405.1 NWSH01002259 PCG68782.1 KY707225 AVF19667.1 SOQ35228.1 GENK01000077 JAV45836.1 GCVX01000220 JAI18010.1 KY283622 AST36281.1 KT588118 AOE48028.1 MG816614 AXF48799.1 KR935716 ALM26208.1 KM892406 AJF23821.1 KM597231 AIT69908.1 KM678332 AJE25905.1 GDKB01000055 JAP38441.1 MG816620 AXF48805.1 KC960481 AGY14592.1 KR935725 ALM26217.1 KR935757 ALM26249.1 KF768702 AIG51896.1 KM892399 AJF23814.1 KY825770 ARF06964.1 PCG68780.1 PCG68779.1 KC960482 AGY14593.1 LC002732 BAR43480.1 KY283619 AST36278.1 KY283678 AST36337.1 KY283757 AST36416.1 AB472100 BAH66317.1 ODYU01011979 SOQ58031.1 GEDO01000059 JAP88567.1 KX084488 ARO76443.1 KF768676 AIG51870.1 LN885119 CUQ99408.1 KT588150 AOE48060.1 KY707218 AVF19660.1 KY283628 AST36287.1 PCG68781.1 KF768677 AIG51871.1 KF768675 AIG51869.1 KF487691 AII01089.1 MF625594 AXY83421.1 KX585327 ARO70220.1 GENK01000081 JAV45832.1 LN885130 CUQ99419.1 KF487654 AII01052.1 LN885104 CUQ99393.1 BK005928 DAA05978.1 ODYU01008612 SOQ52344.1 NWSH01002695 PCG67665.1 KX655966 AOG12915.1 GEDO01000025 JAP88601.1 KX084495 ARO76450.1 AB472098 BAH66315.1 KQ460622 KPJ13512.1 KX655957 AOG12906.1 AGBW02008859 OWR52357.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
A7E3G2
Q0EEG1
A0A076E7L0
A0A2K8GKX5
A0A076E611
E1ACG2
+ More
A0A0P1J400 A0A2K8GL46 A0A1B3B743 A0A2L1TG43 A0A2A4J5Z4 A0A2H1V355 A0A345BEX6 M1QVM8 A0A1V1WBU5 A0A223HD69 Q6A1J3 A0A0B5A1G3 A0A0B5CVM5 A0A2A4J6Z3 A0A097ITW0 A0A075T650 A0A097IYK2 A0A223HD63 H9A5Q2 A0A1B3B785 A0A223HDE8 A0A0K8TU59 A0A223HDG0 A0A2A4JBI2 A0A2L1TG66 A0A2H1V308 A0A1V1WC43 A0A0K8TVB3 A0A223HD37 A0A1B3B736 A0A345BEX5 A0A0S1TPQ8 A0A0B5GIB9 A0A097ITX4 A0A0B5CN09 A0A0V0J201 A0A345BEY1 U5NFU1 A0A0S1TPH0 A0A0S1TPK3 A0A075TD77 A0A0B5GUX0 A0A1W5T543 A0A2A4JBH1 A0A2A4JAB0 U5NJ19 A0A0E4B3W6 A0A223HD26 A0A223HD79 A0A223HDF4 C4B7V1 A0A2H1WY92 A0A146JW15 A0A1X9PBW3 A0A075T629 A0A0P1IVT1 A0A1B3B764 A0A2L1TG92 A0A223HD28 A0A2A4J9Y6 A0A075TD59 A0A075T6W5 A0A076E9B5 A0A3Q8HD97 A0A2K8GKR3 A0A1V1WBU9 A0A0P1IW73 A0A076E7I6 A0A0P1IVQ9 A7E3G3 A0A2H1WIS2 A0A2A4J6F5 A0A1B3P5N2 A0A146JYX6 A0A1Y9TJJ8 C4B7U9 A0A0N1PGA1 A0A1B3P5L3 A0A212FF61
A0A0P1J400 A0A2K8GL46 A0A1B3B743 A0A2L1TG43 A0A2A4J5Z4 A0A2H1V355 A0A345BEX6 M1QVM8 A0A1V1WBU5 A0A223HD69 Q6A1J3 A0A0B5A1G3 A0A0B5CVM5 A0A2A4J6Z3 A0A097ITW0 A0A075T650 A0A097IYK2 A0A223HD63 H9A5Q2 A0A1B3B785 A0A223HDE8 A0A0K8TU59 A0A223HDG0 A0A2A4JBI2 A0A2L1TG66 A0A2H1V308 A0A1V1WC43 A0A0K8TVB3 A0A223HD37 A0A1B3B736 A0A345BEX5 A0A0S1TPQ8 A0A0B5GIB9 A0A097ITX4 A0A0B5CN09 A0A0V0J201 A0A345BEY1 U5NFU1 A0A0S1TPH0 A0A0S1TPK3 A0A075TD77 A0A0B5GUX0 A0A1W5T543 A0A2A4JBH1 A0A2A4JAB0 U5NJ19 A0A0E4B3W6 A0A223HD26 A0A223HD79 A0A223HDF4 C4B7V1 A0A2H1WY92 A0A146JW15 A0A1X9PBW3 A0A075T629 A0A0P1IVT1 A0A1B3B764 A0A2L1TG92 A0A223HD28 A0A2A4J9Y6 A0A075TD59 A0A075T6W5 A0A076E9B5 A0A3Q8HD97 A0A2K8GKR3 A0A1V1WBU9 A0A0P1IW73 A0A076E7I6 A0A0P1IVQ9 A7E3G3 A0A2H1WIS2 A0A2A4J6F5 A0A1B3P5N2 A0A146JYX6 A0A1Y9TJJ8 C4B7U9 A0A0N1PGA1 A0A1B3P5L3 A0A212FF61
Ontologies
GO
PANTHER
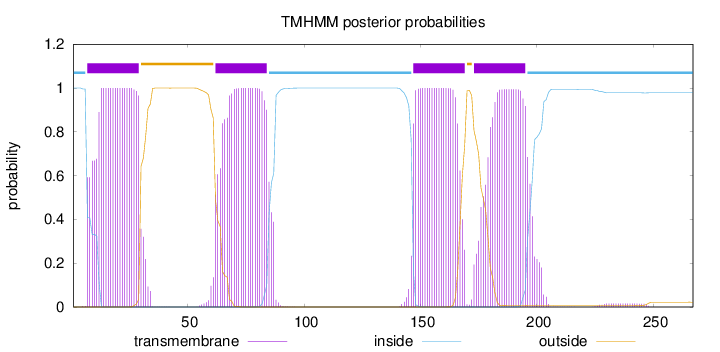
Topology
Subcellular location
Cell membrane
Length:
267
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
87.56819
Exp number, first 60 AAs:
22.28475
Total prob of N-in:
0.99987
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 61
TMhelix
62 - 84
inside
85 - 146
TMhelix
147 - 169
outside
170 - 172
TMhelix
173 - 195
inside
196 - 267
Population Genetic Test Statistics
Pi
232.62772
Theta
178.398738
Tajima's D
0.44126
CLR
0
CSRT
0.503074846257687
Interpretation
Uncertain
