Gene
KWMTBOMO05363
Annotation
PREDICTED:_palmitoyltransferase_ZDHHC6-like_[Bombyx_mori]
Full name
Palmitoyltransferase
Location in the cell
PlasmaMembrane Reliability : 3.383
Sequence
CDS
ATGGTTCTCTCTCCGTTTCGACGATTATTGCACTGGGGTCCCATATCGGTTTTAGGTATAATAAAATTAATAACATGGGCCATGGTCCACATGCTGGGAATGTGGTGGCCCCCTCAAACTTCATTCTATGCATCGCTTCATGCAGCAATGTTCCTGTCATTTGCTGCGGGAACATTATATTATTTCTTGCAATCATTACTTGAGGGACCTGGGTTCGTGCCTTTGGGATGGCAGCCTGACAAAGAAGAGGATGCCCAATATCTACAATTTTGCACAATTTGTAAAGGATACAAGGCTCCACGGTCACACCACTGTAGGAAATGTGGCTTGTGCGTTTTGAAGATGGACCATCATTGCCCGTGGATAAATTGTTGCGTTGGTCACGCTAACCATGGGTATTTCACTACTTTTCTAATGTTTGCAGTTCTTGGCTGCTTCCATGCCTCTGTAGTATTATCCATTTGTATGTATCATGCGATAAATCGGATATGGTACATCCACCACGGCTCCGGTGACGAGCCCATGATCTACGTCACTTTGACTACTTTACTGCTGACCCTACTCTCAATAGGGATGGCTGTTGGTGTAGTACTCGCTGTTGGTACATTACTTTATTTGCAGATTCGAGGCATACTTCGTAATCAGACCACGATCGAGGACTGGATCATGGAAAAAGCTGCAGGTAGACGAGAAGAACAGGGTCTGCCTCCGTTCGTGTTCCCTTACGACGTGGGCTGGAAGAGGAATATAGCTTCGGTGCTGTCTCGGAGTAGAGGGGATGGAATCCGGTGGCCCTTGAAGGATGGCTGTGGCCAGTATGATCTGACCCGCGAGCAGCAGGCGCAGAAGGCGGACAAGCGCGTGCGGGCGCGCCTGTACGTGTGCCGCGCGGCCTACTCCGGGCGCTGGTGTCCGCTGGCGCAGCACCCCCGCGCCGCGCTGGCGCCGCCCTGCACGGACGAGCCGCGCCTGCCGCTGGCGCCGGGCGACGCGCTGCGGGCCACGCGCCACCGCCGCCACTGGCTGTTCGGCGAGCGGCTGGCGGGCGCCGGCGGGGGCGAGGGCGGGGCGCGGGGGCCGCGGGGCTGGTTCCCGCGCAACTGCGTGCTGCCCGAGCGCGACCCCGCCAAAGCCAAGCCCAACTAG
Protein
MVLSPFRRLLHWGPISVLGIIKLITWAMVHMLGMWWPPQTSFYASLHAAMFLSFAAGTLYYFLQSLLEGPGFVPLGWQPDKEEDAQYLQFCTICKGYKAPRSHHCRKCGLCVLKMDHHCPWINCCVGHANHGYFTTFLMFAVLGCFHASVVLSICMYHAINRIWYIHHGSGDEPMIYVTLTTLLLTLLSIGMAVGVVLAVGTLLYLQIRGILRNQTTIEDWIMEKAAGRREEQGLPPFVFPYDVGWKRNIASVLSRSRGDGIRWPLKDGCGQYDLTREQQAQKADKRVRARLYVCRAAYSGRWCPLAQHPRAALAPPCTDEPRLPLAPGDALRATRHRRHWLFGERLAGAGGGEGGARGPRGWFPRNCVLPERDPAKAKPN
Summary
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Similarity
Belongs to the DHHC palmitoyltransferase family.
Uniprot
A0A2A4J9X6
A0A2H1V670
A0A194QIQ5
A0A212EUT6
A0A3S2LFW4
A0A0N0PBY0
+ More
A0A2A4JBG1 D6WBK9 A0A023F1K0 A0A069DTJ1 A0A224XD47 A0A0V0GC15 A0A1B6DZI2 A0A182Y4U2 A0A182NI16 A0A182RM21 A0A0P4VIP1 A0A0A9Z6P4 A0A1L8DY07 A0A0P4WCK0 A0A1L8DXY8 A0A182QUP0 A0A2M4CYP7 W5JWP0 B4JTP9 A0A182VV46 B4K872 A0A1A9XVM3 A0A1A9ZWV3 D3TPN4 A0A1B0BKZ0 A0A1Y1K8V9 A0A0M4EQG2 A0A1B6LI42 A0A2J7Q2E8 B4LY95 A0A1A9VGE0 W8B3X2 A0A084W9N7 A0A0A1WK71 A0A182WY27 A0A182UUH2 Q7PNJ3 A0A0K8U5W1 A0A182HMT6 A0A034WF10 A0A2M4A9G5 A0A1Q3FXU4 A0A1I8N4X9 A0A182PL07 A0A3B0KAA2 A0A1Q3FXX8 A0A182J3I4 B3M2Q2 B4HGY7 A0A2M4BPX2 Q29B32 U5EWU7 B4QSC0 Q9VG80 B4GZ57 A0A1I8Q4F4 A0A1W4UBG8 A0A1B0CF66 A0A087TI87 A0A182K328 A0A1L8DY33 B4PNT1 A0A1L8DXZ5 B3P1B3 A0A232F6K7 N6T8U8 Q16LA8 A0A023ERX0 A0A0L0CEG5 E0VI53 K7J230 E9HB00 A0A1W4X5S6 A0A026WEE2 A0A2T7Q1H5 E2AIW9 A0A0P5J4I7 A0A0C9PRD8 A0A182FMG9 A0A1E1XSD8 B7QJ17 A0A1S3HZZ4 A0A131Z6U1 A0A131XBP9 A0A0Q9WSB1 A0A182GWM7 L7MCF2 A0A1E1XD16 A0A131XVN3 A0A0P5K5P0 A0A2A3E4G4 A0A088ADV8 A0A224ZC37 A0A067RFJ1
A0A2A4JBG1 D6WBK9 A0A023F1K0 A0A069DTJ1 A0A224XD47 A0A0V0GC15 A0A1B6DZI2 A0A182Y4U2 A0A182NI16 A0A182RM21 A0A0P4VIP1 A0A0A9Z6P4 A0A1L8DY07 A0A0P4WCK0 A0A1L8DXY8 A0A182QUP0 A0A2M4CYP7 W5JWP0 B4JTP9 A0A182VV46 B4K872 A0A1A9XVM3 A0A1A9ZWV3 D3TPN4 A0A1B0BKZ0 A0A1Y1K8V9 A0A0M4EQG2 A0A1B6LI42 A0A2J7Q2E8 B4LY95 A0A1A9VGE0 W8B3X2 A0A084W9N7 A0A0A1WK71 A0A182WY27 A0A182UUH2 Q7PNJ3 A0A0K8U5W1 A0A182HMT6 A0A034WF10 A0A2M4A9G5 A0A1Q3FXU4 A0A1I8N4X9 A0A182PL07 A0A3B0KAA2 A0A1Q3FXX8 A0A182J3I4 B3M2Q2 B4HGY7 A0A2M4BPX2 Q29B32 U5EWU7 B4QSC0 Q9VG80 B4GZ57 A0A1I8Q4F4 A0A1W4UBG8 A0A1B0CF66 A0A087TI87 A0A182K328 A0A1L8DY33 B4PNT1 A0A1L8DXZ5 B3P1B3 A0A232F6K7 N6T8U8 Q16LA8 A0A023ERX0 A0A0L0CEG5 E0VI53 K7J230 E9HB00 A0A1W4X5S6 A0A026WEE2 A0A2T7Q1H5 E2AIW9 A0A0P5J4I7 A0A0C9PRD8 A0A182FMG9 A0A1E1XSD8 B7QJ17 A0A1S3HZZ4 A0A131Z6U1 A0A131XBP9 A0A0Q9WSB1 A0A182GWM7 L7MCF2 A0A1E1XD16 A0A131XVN3 A0A0P5K5P0 A0A2A3E4G4 A0A088ADV8 A0A224ZC37 A0A067RFJ1
EC Number
2.3.1.225
Pubmed
26354079
22118469
18362917
19820115
25474469
26334808
+ More
25244985 27129103 25401762 20920257 23761445 17994087 20353571 28004739 24495485 24438588 25830018 12364791 14747013 17210077 25348373 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28648823 23537049 17510324 24945155 26108605 20566863 20075255 21292972 24508170 30249741 20798317 29209593 26830274 28049606 26483478 25576852 28503490 28797301 24845553
25244985 27129103 25401762 20920257 23761445 17994087 20353571 28004739 24495485 24438588 25830018 12364791 14747013 17210077 25348373 25315136 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28648823 23537049 17510324 24945155 26108605 20566863 20075255 21292972 24508170 30249741 20798317 29209593 26830274 28049606 26483478 25576852 28503490 28797301 24845553
EMBL
NWSH01002259
PCG68771.1
ODYU01000898
SOQ36350.1
KQ458714
KPJ05438.1
+ More
AGBW02012297 OWR45252.1 RSAL01000148 RVE45830.1 KQ460864 KPJ11805.1 PCG68770.1 KQ971311 EEZ99000.2 GBBI01003539 JAC15173.1 GBGD01001808 JAC87081.1 GFTR01006050 JAW10376.1 GECL01001197 JAP04927.1 GEDC01006206 JAS31092.1 GDKW01001864 JAI54731.1 GBHO01002667 GBRD01002144 JAG40937.1 JAG63677.1 GFDF01002756 JAV11328.1 GDRN01063555 JAI64961.1 GFDF01002757 JAV11327.1 AXCN02000387 GGFL01006239 MBW70417.1 ADMH02000121 ETN67680.1 CH916374 EDV91478.1 CH933806 EDW15426.1 CCAG010020908 EZ423386 ADD19662.1 JXJN01016128 GEZM01095997 JAV55217.1 CP012526 ALC46572.1 GEBQ01016601 JAT23376.1 NEVH01019371 PNF22756.1 CH940650 EDW67983.1 GAMC01010720 JAB95835.1 ATLV01021848 KE525324 KFB46931.1 GBXI01014843 JAC99448.1 AAAB01008963 EAA12087.6 GDHF01030275 JAI22039.1 APCN01004903 GAKP01006584 JAC52368.1 GGFK01004049 MBW37370.1 GFDL01002671 JAV32374.1 OUUW01000005 SPP80488.1 GFDL01002689 JAV32356.1 CH902617 EDV42373.1 CH480815 EDW42458.1 GGFJ01005830 MBW54971.1 CM000070 EAL27167.1 GANO01002807 JAB57064.1 CM000364 EDX13216.1 AE014297 BT082042 AAF54805.1 ACO72879.1 CH479198 EDW28075.1 AJWK01009735 AJWK01009736 KK115334 KFM64826.1 GFDF01002760 JAV11324.1 CM000160 EDW97096.1 GFDF01002761 JAV11323.1 CH954181 EDV49372.1 NNAY01000882 OXU26078.1 APGK01039372 KB740970 ENN76649.1 CH477912 EAT35099.1 GAPW01001640 JAC11958.1 JRES01000501 KNC30641.1 DS235184 EEB13059.1 GL732613 EFX71118.1 KK107293 QOIP01000005 EZA53399.1 RLU22667.1 PZQS01000001 PVD39528.1 GL439905 EFN66648.1 GDIQ01207161 JAK44564.1 GBYB01003838 JAG73605.1 GFAA01001216 JAU02219.1 ABJB010303371 ABJB010613420 DS949079 EEC18839.1 GEDV01001859 JAP86698.1 GEFH01005021 JAP63560.1 KRF83598.1 JXUM01093682 KQ564073 KXJ72828.1 GACK01003384 JAA61650.1 GFAC01002187 JAT97001.1 GEFM01005528 JAP70268.1 GDIQ01190468 GDIQ01162778 GDIP01054921 LRGB01000687 JAK61257.1 JAM48794.1 KZS16825.1 KZ288416 PBC26049.1 GFPF01013446 MAA24592.1 KK852667 KDR18933.1
AGBW02012297 OWR45252.1 RSAL01000148 RVE45830.1 KQ460864 KPJ11805.1 PCG68770.1 KQ971311 EEZ99000.2 GBBI01003539 JAC15173.1 GBGD01001808 JAC87081.1 GFTR01006050 JAW10376.1 GECL01001197 JAP04927.1 GEDC01006206 JAS31092.1 GDKW01001864 JAI54731.1 GBHO01002667 GBRD01002144 JAG40937.1 JAG63677.1 GFDF01002756 JAV11328.1 GDRN01063555 JAI64961.1 GFDF01002757 JAV11327.1 AXCN02000387 GGFL01006239 MBW70417.1 ADMH02000121 ETN67680.1 CH916374 EDV91478.1 CH933806 EDW15426.1 CCAG010020908 EZ423386 ADD19662.1 JXJN01016128 GEZM01095997 JAV55217.1 CP012526 ALC46572.1 GEBQ01016601 JAT23376.1 NEVH01019371 PNF22756.1 CH940650 EDW67983.1 GAMC01010720 JAB95835.1 ATLV01021848 KE525324 KFB46931.1 GBXI01014843 JAC99448.1 AAAB01008963 EAA12087.6 GDHF01030275 JAI22039.1 APCN01004903 GAKP01006584 JAC52368.1 GGFK01004049 MBW37370.1 GFDL01002671 JAV32374.1 OUUW01000005 SPP80488.1 GFDL01002689 JAV32356.1 CH902617 EDV42373.1 CH480815 EDW42458.1 GGFJ01005830 MBW54971.1 CM000070 EAL27167.1 GANO01002807 JAB57064.1 CM000364 EDX13216.1 AE014297 BT082042 AAF54805.1 ACO72879.1 CH479198 EDW28075.1 AJWK01009735 AJWK01009736 KK115334 KFM64826.1 GFDF01002760 JAV11324.1 CM000160 EDW97096.1 GFDF01002761 JAV11323.1 CH954181 EDV49372.1 NNAY01000882 OXU26078.1 APGK01039372 KB740970 ENN76649.1 CH477912 EAT35099.1 GAPW01001640 JAC11958.1 JRES01000501 KNC30641.1 DS235184 EEB13059.1 GL732613 EFX71118.1 KK107293 QOIP01000005 EZA53399.1 RLU22667.1 PZQS01000001 PVD39528.1 GL439905 EFN66648.1 GDIQ01207161 JAK44564.1 GBYB01003838 JAG73605.1 GFAA01001216 JAU02219.1 ABJB010303371 ABJB010613420 DS949079 EEC18839.1 GEDV01001859 JAP86698.1 GEFH01005021 JAP63560.1 KRF83598.1 JXUM01093682 KQ564073 KXJ72828.1 GACK01003384 JAA61650.1 GFAC01002187 JAT97001.1 GEFM01005528 JAP70268.1 GDIQ01190468 GDIQ01162778 GDIP01054921 LRGB01000687 JAK61257.1 JAM48794.1 KZS16825.1 KZ288416 PBC26049.1 GFPF01013446 MAA24592.1 KK852667 KDR18933.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000283053
UP000053240
UP000007266
+ More
UP000076408 UP000075884 UP000075900 UP000075886 UP000000673 UP000001070 UP000075920 UP000009192 UP000092443 UP000092445 UP000092444 UP000092460 UP000092553 UP000235965 UP000008792 UP000078200 UP000030765 UP000076407 UP000075903 UP000007062 UP000075840 UP000095301 UP000075885 UP000268350 UP000075880 UP000007801 UP000001292 UP000001819 UP000000304 UP000000803 UP000008744 UP000095300 UP000192221 UP000092461 UP000054359 UP000075881 UP000002282 UP000008711 UP000215335 UP000019118 UP000008820 UP000037069 UP000009046 UP000002358 UP000000305 UP000192223 UP000053097 UP000279307 UP000245119 UP000000311 UP000069272 UP000001555 UP000085678 UP000069940 UP000249989 UP000076858 UP000242457 UP000005203 UP000027135
UP000076408 UP000075884 UP000075900 UP000075886 UP000000673 UP000001070 UP000075920 UP000009192 UP000092443 UP000092445 UP000092444 UP000092460 UP000092553 UP000235965 UP000008792 UP000078200 UP000030765 UP000076407 UP000075903 UP000007062 UP000075840 UP000095301 UP000075885 UP000268350 UP000075880 UP000007801 UP000001292 UP000001819 UP000000304 UP000000803 UP000008744 UP000095300 UP000192221 UP000092461 UP000054359 UP000075881 UP000002282 UP000008711 UP000215335 UP000019118 UP000008820 UP000037069 UP000009046 UP000002358 UP000000305 UP000192223 UP000053097 UP000279307 UP000245119 UP000000311 UP000069272 UP000001555 UP000085678 UP000069940 UP000249989 UP000076858 UP000242457 UP000005203 UP000027135
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J9X6
A0A2H1V670
A0A194QIQ5
A0A212EUT6
A0A3S2LFW4
A0A0N0PBY0
+ More
A0A2A4JBG1 D6WBK9 A0A023F1K0 A0A069DTJ1 A0A224XD47 A0A0V0GC15 A0A1B6DZI2 A0A182Y4U2 A0A182NI16 A0A182RM21 A0A0P4VIP1 A0A0A9Z6P4 A0A1L8DY07 A0A0P4WCK0 A0A1L8DXY8 A0A182QUP0 A0A2M4CYP7 W5JWP0 B4JTP9 A0A182VV46 B4K872 A0A1A9XVM3 A0A1A9ZWV3 D3TPN4 A0A1B0BKZ0 A0A1Y1K8V9 A0A0M4EQG2 A0A1B6LI42 A0A2J7Q2E8 B4LY95 A0A1A9VGE0 W8B3X2 A0A084W9N7 A0A0A1WK71 A0A182WY27 A0A182UUH2 Q7PNJ3 A0A0K8U5W1 A0A182HMT6 A0A034WF10 A0A2M4A9G5 A0A1Q3FXU4 A0A1I8N4X9 A0A182PL07 A0A3B0KAA2 A0A1Q3FXX8 A0A182J3I4 B3M2Q2 B4HGY7 A0A2M4BPX2 Q29B32 U5EWU7 B4QSC0 Q9VG80 B4GZ57 A0A1I8Q4F4 A0A1W4UBG8 A0A1B0CF66 A0A087TI87 A0A182K328 A0A1L8DY33 B4PNT1 A0A1L8DXZ5 B3P1B3 A0A232F6K7 N6T8U8 Q16LA8 A0A023ERX0 A0A0L0CEG5 E0VI53 K7J230 E9HB00 A0A1W4X5S6 A0A026WEE2 A0A2T7Q1H5 E2AIW9 A0A0P5J4I7 A0A0C9PRD8 A0A182FMG9 A0A1E1XSD8 B7QJ17 A0A1S3HZZ4 A0A131Z6U1 A0A131XBP9 A0A0Q9WSB1 A0A182GWM7 L7MCF2 A0A1E1XD16 A0A131XVN3 A0A0P5K5P0 A0A2A3E4G4 A0A088ADV8 A0A224ZC37 A0A067RFJ1
A0A2A4JBG1 D6WBK9 A0A023F1K0 A0A069DTJ1 A0A224XD47 A0A0V0GC15 A0A1B6DZI2 A0A182Y4U2 A0A182NI16 A0A182RM21 A0A0P4VIP1 A0A0A9Z6P4 A0A1L8DY07 A0A0P4WCK0 A0A1L8DXY8 A0A182QUP0 A0A2M4CYP7 W5JWP0 B4JTP9 A0A182VV46 B4K872 A0A1A9XVM3 A0A1A9ZWV3 D3TPN4 A0A1B0BKZ0 A0A1Y1K8V9 A0A0M4EQG2 A0A1B6LI42 A0A2J7Q2E8 B4LY95 A0A1A9VGE0 W8B3X2 A0A084W9N7 A0A0A1WK71 A0A182WY27 A0A182UUH2 Q7PNJ3 A0A0K8U5W1 A0A182HMT6 A0A034WF10 A0A2M4A9G5 A0A1Q3FXU4 A0A1I8N4X9 A0A182PL07 A0A3B0KAA2 A0A1Q3FXX8 A0A182J3I4 B3M2Q2 B4HGY7 A0A2M4BPX2 Q29B32 U5EWU7 B4QSC0 Q9VG80 B4GZ57 A0A1I8Q4F4 A0A1W4UBG8 A0A1B0CF66 A0A087TI87 A0A182K328 A0A1L8DY33 B4PNT1 A0A1L8DXZ5 B3P1B3 A0A232F6K7 N6T8U8 Q16LA8 A0A023ERX0 A0A0L0CEG5 E0VI53 K7J230 E9HB00 A0A1W4X5S6 A0A026WEE2 A0A2T7Q1H5 E2AIW9 A0A0P5J4I7 A0A0C9PRD8 A0A182FMG9 A0A1E1XSD8 B7QJ17 A0A1S3HZZ4 A0A131Z6U1 A0A131XBP9 A0A0Q9WSB1 A0A182GWM7 L7MCF2 A0A1E1XD16 A0A131XVN3 A0A0P5K5P0 A0A2A3E4G4 A0A088ADV8 A0A224ZC37 A0A067RFJ1
PDB
6BMS
E-value=3.57619e-13,
Score=181
Ontologies
GO
PANTHER
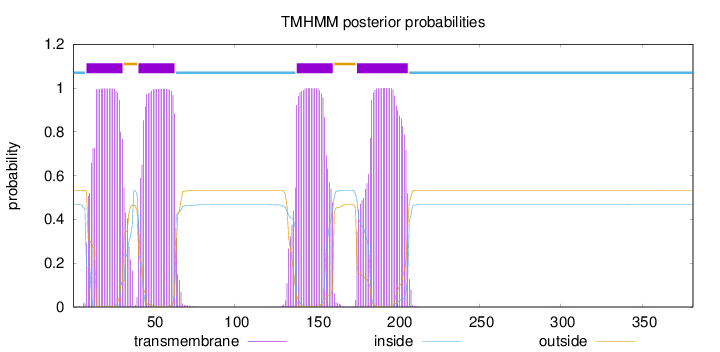
Topology
Length:
381
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
92.8292599999999
Exp number, first 60 AAs:
40.25718
Total prob of N-in:
0.46809
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 40
TMhelix
41 - 63
inside
64 - 137
TMhelix
138 - 160
outside
161 - 174
TMhelix
175 - 206
inside
207 - 381
Population Genetic Test Statistics
Pi
308.120002
Theta
181.034584
Tajima's D
2.179673
CLR
0.101356
CSRT
0.912304384780761
Interpretation
Uncertain
