Pre Gene Modal
BGIBMGA002379
Annotation
PREDICTED:_dynactin_subunit_4_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.696
Sequence
CDS
ATGGCATATTTAACTCAACCTGATTATGTGAGATACATTTGTACTTGCGGTCAACTGAAACCAATCACTAGTTTATACTTCTGTCGGCATTGTTTGAAAATACGTTGCGGTTTTTGTATTTGTCATGAGGTCGACTCACATTATTGTCCGAACTGCCTCGAGAATATGCCATCATCAGAAGCCAGATTAAAGAAGAACCGTTGCAGCAGCTGCTTCGATTGTCCCAGTTGCTACCACACTTTGTCCACCAGAGCTAGCCTGGTCCAACCTCGGCAGCAAGAAGGTGCTGGAGATGGGAAAGTAGAAAACAAACATCAAAAAAAGATGTACTATTTAGCTTGTTTCAATTGTAGATGGACATCTAGAGATGTGGGAATTCCTGATCAGCCTGTTGCATCAGGTGGATGGCCAGAGAGACCAAATCCATTTGCAACACAAATCAATCAACTCTTAGAATATTACAAGGCAATAGCATTACAAGAGAAACAGGAGAAAATTGAAAAAGAAAGAAAAAAGTTTGCCATCAGAGGGAAATACCTAACACTCACTGATAAAACGGGCTTAACTGGAGCTATGGCAAGGAACATAGCAGGACTTCCATCTTCCGACTCTGGGTCATCGTCCATTATCGCCAAATTCGTTCCAAGCCAAGCCAGTGCAGAAGTTGAGGAATTGCCCGAAGATTATTTCACTAAAGAAGTCAACTTGAATCAGATAACCAGTATGCCTCAACGCCTTCTATCACCGGAGTGGCAGCCTACTGATGTTTCAAGACTGCATCCAATAGCTAAACTATTGAGTGTAAAACGTAGCCAACGGTGCAGGGAGTGTGACCACAATCTGACTAAACCAGAGTATAATCCGGGCTCTATAAAATTCAAGATTGAGTTACTGGCTTTTTATCACATACCCGAAGTGAAAATAATATCATACGAACCCATGAAGGCAGGTGAGAATTCAACTGTGTTGTTGAAACTGAGCAATCCAACTGCTCACGAAATGACATTATGCCTGATGAATCCTGAAGATATACCGCAAGTGGACAATGAGGAAGAGCAACTTGAAAATTCACTTGAGAAATCACTCACTTTGGACAAGGAGTCCGCGGCGTGCAAGCCGGCCGCCGCGCCCGCTTGGCGCCGCGCCGCCGCGCCGTGCGTCCGCGCCGTCGGGCCGCCCGTGCGCGTCTCCGTGCCGCAGCGCGACGACGCCGCCGAGTACGACGACGACGCCACGCACGACGCTAACGATGTAATCGTGTGGCGAAAGGCGAACAAGCTGGGCGTCCGTCTGCAGGTGGCGACGGAGGCGGCGGCGTCGCGGGGCGGCGCGGCGCTGCTGGCGCTGGCCCTCCAGTACTCGTACCGCAATACGGTGCCGCCCGCCGCCAACGCGCCGAACCAGAGGAATCATATTCTGTACACGACGGCGTTGCTCTCACTGGGAAACGTAGCTTAA
Protein
MAYLTQPDYVRYICTCGQLKPITSLYFCRHCLKIRCGFCICHEVDSHYCPNCLENMPSSEARLKKNRCSSCFDCPSCYHTLSTRASLVQPRQQEGAGDGKVENKHQKKMYYLACFNCRWTSRDVGIPDQPVASGGWPERPNPFATQINQLLEYYKAIALQEKQEKIEKERKKFAIRGKYLTLTDKTGLTGAMARNIAGLPSSDSGSSSIIAKFVPSQASAEVEELPEDYFTKEVNLNQITSMPQRLLSPEWQPTDVSRLHPIAKLLSVKRSQRCRECDHNLTKPEYNPGSIKFKIELLAFYHIPEVKIISYEPMKAGENSTVLLKLSNPTAHEMTLCLMNPEDIPQVDNEEEQLENSLEKSLTLDKESAACKPAAAPAWRRAAAPCVRAVGPPVRVSVPQRDDAAEYDDDATHDANDVIVWRKANKLGVRLQVATEAAASRGGAALLALALQYSYRNTVPPAANAPNQRNHILYTTALLSLGNVA
Summary
Uniprot
H9IYP5
A0A2A4IV45
A0A3S2NAG8
A0A0N1IAF4
A0A194QPN2
A0A212EUT8
+ More
A0A2H1V6E3 A0A2J7PY88 A0A1Y1JZ50 D6X4D7 A0A3R7M8D9 A0A0P4VW45 T1IG42 A0A069DZ40 K7J4L4 A0A151I3X3 E2BDW5 A0A0C9R983 A0A0V0GAT3 A0A1B6JFK8 A0A1B6DNW3 A0A0P4W2P9 A0A224XBS8 A0A1W4WQT5 A0A1B6FV03 B4LJG9 A0A0N0BC75 A0A026WAB7 A0A158NYM2 E9JDH4 F4WXY9 T1GM44 A0A0K8TMS8 A0A1J1HKN3 A0A067QMH3 A0A1Q3G1N4 J7G8Y8 A0A1Q3G1Q4 A0A1Q3G1R1 Q17AJ7 A0A1Q3G1V0 A0A0A9X5U0 A0A1Q3G1K6 A0A0A1XLE9 A0A154NY09 A0A0L7RH23 W8CCU5 A0A0K8TWA1 A0A084VC99 B4HR53 Q7K130 A0A1B0GN13 A0A034W8J4 A0A182IWX8 B4J7Z7 A0A0M4EHH9 B3N9V6 B4N653 A0A0K8RIH6 V5HDN0 B4QEQ8 A0A182NED6 B4P1T3 A0A1B0CAC1 A0A0K8VVY5 A0A3B0J2Z4 B4KPR2 Q290Q3 A0A1B0CK06 A0A1W4WB48 B3MHS5 A0A182QQ57 B4GBE3 Q17AJ6 A0A1Z5KWK4 A0A088ADL4 A0A182URW3 Q7PNK5 A0A182RKC8 A0A182JXB4 A0A182XNK4 A0A1L8E5D8 A0A1B0GGF6 A0A1B0AT09 A0A182HMV7 A0A224YXK6 A0A131YPU0 A0A1A9YGV5 A0A131XNP4 U4TYN6 A0A182TUX7 A0A2M4AME9 A0A1I8N7K1 B7PEE3 T1JF52 D3TQ10
A0A2H1V6E3 A0A2J7PY88 A0A1Y1JZ50 D6X4D7 A0A3R7M8D9 A0A0P4VW45 T1IG42 A0A069DZ40 K7J4L4 A0A151I3X3 E2BDW5 A0A0C9R983 A0A0V0GAT3 A0A1B6JFK8 A0A1B6DNW3 A0A0P4W2P9 A0A224XBS8 A0A1W4WQT5 A0A1B6FV03 B4LJG9 A0A0N0BC75 A0A026WAB7 A0A158NYM2 E9JDH4 F4WXY9 T1GM44 A0A0K8TMS8 A0A1J1HKN3 A0A067QMH3 A0A1Q3G1N4 J7G8Y8 A0A1Q3G1Q4 A0A1Q3G1R1 Q17AJ7 A0A1Q3G1V0 A0A0A9X5U0 A0A1Q3G1K6 A0A0A1XLE9 A0A154NY09 A0A0L7RH23 W8CCU5 A0A0K8TWA1 A0A084VC99 B4HR53 Q7K130 A0A1B0GN13 A0A034W8J4 A0A182IWX8 B4J7Z7 A0A0M4EHH9 B3N9V6 B4N653 A0A0K8RIH6 V5HDN0 B4QEQ8 A0A182NED6 B4P1T3 A0A1B0CAC1 A0A0K8VVY5 A0A3B0J2Z4 B4KPR2 Q290Q3 A0A1B0CK06 A0A1W4WB48 B3MHS5 A0A182QQ57 B4GBE3 Q17AJ6 A0A1Z5KWK4 A0A088ADL4 A0A182URW3 Q7PNK5 A0A182RKC8 A0A182JXB4 A0A182XNK4 A0A1L8E5D8 A0A1B0GGF6 A0A1B0AT09 A0A182HMV7 A0A224YXK6 A0A131YPU0 A0A1A9YGV5 A0A131XNP4 U4TYN6 A0A182TUX7 A0A2M4AME9 A0A1I8N7K1 B7PEE3 T1JF52 D3TQ10
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
27129103 26334808 20075255 20798317 17994087 24508170 21347285 21282665 21719571 26369729 24845553 17510324 25401762 26823975 25830018 24495485 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25765539 22936249 17550304 15632085 28528879 12364791 14747013 17210077 28797301 26830274 28049606 23537049 25315136 20353571
27129103 26334808 20075255 20798317 17994087 24508170 21347285 21282665 21719571 26369729 24845553 17510324 25401762 26823975 25830018 24495485 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 25765539 22936249 17550304 15632085 28528879 12364791 14747013 17210077 28797301 26830274 28049606 23537049 25315136 20353571
EMBL
BABH01011158
BABH01011159
NWSH01007004
PCG63154.1
RSAL01000148
RVE45833.1
+ More
KQ459934 KPJ19264.1 KQ458714 KPJ05441.1 AGBW02012297 OWR45255.1 ODYU01000898 SOQ36346.1 NEVH01020850 PNF21260.1 GEZM01099075 JAV53511.1 KQ971410 EEZ97538.1 QCYY01001738 ROT75734.1 GDKW01000281 JAI56314.1 ACPB03002684 GBGD01001305 JAC87584.1 AAZX01002331 KQ976471 KYM84027.1 GL447693 EFN86091.1 GBYB01009432 JAG79199.1 GECL01001151 JAP04973.1 GECU01009747 JAS97959.1 GEDC01016284 GEDC01009922 GEDC01008476 JAS21014.1 JAS27376.1 JAS28822.1 GDRN01089647 JAI60618.1 GFTR01006601 JAW09825.1 GECZ01019711 GECZ01015733 GECZ01011820 GECZ01011367 JAS50058.1 JAS54036.1 JAS57949.1 JAS58402.1 CH940648 EDW61537.1 KQ435944 KOX68241.1 KK107295 EZA52995.1 ADTU01004054 GL771866 EFZ09136.1 GL888434 EGI60925.1 CAQQ02199958 GDAI01002152 JAI15451.1 CVRI01000006 CRK88090.1 KK853245 KDR09402.1 GFDL01001307 JAV33738.1 JX101463 JX101464 KZ288206 AFP58805.1 PBC33088.1 GFDL01001323 JAV33722.1 GFDL01001315 JAV33730.1 CH477333 EAT43296.1 GFDL01001294 JAV33751.1 GBHO01029456 GBRD01015346 GDHC01010571 JAG14148.1 JAG50480.1 JAQ08058.1 GFDL01001361 JAV33684.1 GBXI01002909 JAD11383.1 KQ434773 KZC03974.1 KQ414596 KOC70001.1 GAMC01003086 JAC03470.1 GDHF01033959 JAI18355.1 ATLV01010627 KE524589 KFB35593.1 CH480816 EDW46796.1 AE013599 AY069420 AAF59211.1 AAL39565.1 AJVK01027889 GAKP01008335 GAKP01008334 JAC50617.1 CH916367 EDW02227.1 CP012524 ALC42022.1 CH954177 EDV59652.1 CH964154 EDW79842.1 GADI01002878 JAA70930.1 GANP01008919 JAB75549.1 CM000362 CM002911 EDX06054.1 KMY92057.1 CM000157 EDW89219.1 AJWK01003628 GDHF01009278 JAI43036.1 OUUW01000001 SPP73572.1 CH933808 EDW09172.1 CM000071 EAL25309.2 AJWK01015591 CH902619 EDV36912.1 AXCN02000860 CH479181 EDW32245.1 EAT43297.1 GFJQ02007474 JAV99495.1 AAAB01008963 EAA12052.5 GFDF01000166 JAV13918.1 CCAG010015359 JXJN01003090 APCN01004916 GFPF01007416 MAA18562.1 GEDV01007580 JAP80977.1 GEFH01000851 JAP67730.1 KB631665 ERL85138.1 GGFK01008581 MBW41902.1 ABJB010110598 ABJB010848507 DS695221 EEC04965.1 JH432146 EZ423512 ADD19788.1
KQ459934 KPJ19264.1 KQ458714 KPJ05441.1 AGBW02012297 OWR45255.1 ODYU01000898 SOQ36346.1 NEVH01020850 PNF21260.1 GEZM01099075 JAV53511.1 KQ971410 EEZ97538.1 QCYY01001738 ROT75734.1 GDKW01000281 JAI56314.1 ACPB03002684 GBGD01001305 JAC87584.1 AAZX01002331 KQ976471 KYM84027.1 GL447693 EFN86091.1 GBYB01009432 JAG79199.1 GECL01001151 JAP04973.1 GECU01009747 JAS97959.1 GEDC01016284 GEDC01009922 GEDC01008476 JAS21014.1 JAS27376.1 JAS28822.1 GDRN01089647 JAI60618.1 GFTR01006601 JAW09825.1 GECZ01019711 GECZ01015733 GECZ01011820 GECZ01011367 JAS50058.1 JAS54036.1 JAS57949.1 JAS58402.1 CH940648 EDW61537.1 KQ435944 KOX68241.1 KK107295 EZA52995.1 ADTU01004054 GL771866 EFZ09136.1 GL888434 EGI60925.1 CAQQ02199958 GDAI01002152 JAI15451.1 CVRI01000006 CRK88090.1 KK853245 KDR09402.1 GFDL01001307 JAV33738.1 JX101463 JX101464 KZ288206 AFP58805.1 PBC33088.1 GFDL01001323 JAV33722.1 GFDL01001315 JAV33730.1 CH477333 EAT43296.1 GFDL01001294 JAV33751.1 GBHO01029456 GBRD01015346 GDHC01010571 JAG14148.1 JAG50480.1 JAQ08058.1 GFDL01001361 JAV33684.1 GBXI01002909 JAD11383.1 KQ434773 KZC03974.1 KQ414596 KOC70001.1 GAMC01003086 JAC03470.1 GDHF01033959 JAI18355.1 ATLV01010627 KE524589 KFB35593.1 CH480816 EDW46796.1 AE013599 AY069420 AAF59211.1 AAL39565.1 AJVK01027889 GAKP01008335 GAKP01008334 JAC50617.1 CH916367 EDW02227.1 CP012524 ALC42022.1 CH954177 EDV59652.1 CH964154 EDW79842.1 GADI01002878 JAA70930.1 GANP01008919 JAB75549.1 CM000362 CM002911 EDX06054.1 KMY92057.1 CM000157 EDW89219.1 AJWK01003628 GDHF01009278 JAI43036.1 OUUW01000001 SPP73572.1 CH933808 EDW09172.1 CM000071 EAL25309.2 AJWK01015591 CH902619 EDV36912.1 AXCN02000860 CH479181 EDW32245.1 EAT43297.1 GFJQ02007474 JAV99495.1 AAAB01008963 EAA12052.5 GFDF01000166 JAV13918.1 CCAG010015359 JXJN01003090 APCN01004916 GFPF01007416 MAA18562.1 GEDV01007580 JAP80977.1 GEFH01000851 JAP67730.1 KB631665 ERL85138.1 GGFK01008581 MBW41902.1 ABJB010110598 ABJB010848507 DS695221 EEC04965.1 JH432146 EZ423512 ADD19788.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000007266 UP000283509 UP000015103 UP000002358 UP000078540 UP000008237 UP000192223 UP000008792 UP000053105 UP000053097 UP000005205 UP000007755 UP000015102 UP000183832 UP000027135 UP000242457 UP000008820 UP000076502 UP000053825 UP000030765 UP000001292 UP000000803 UP000092462 UP000075880 UP000001070 UP000092553 UP000008711 UP000007798 UP000000304 UP000075884 UP000002282 UP000092461 UP000268350 UP000009192 UP000001819 UP000192221 UP000007801 UP000075886 UP000008744 UP000005203 UP000075903 UP000007062 UP000075900 UP000075881 UP000076407 UP000092444 UP000092460 UP000075840 UP000092443 UP000030742 UP000075902 UP000095301 UP000001555
UP000235965 UP000007266 UP000283509 UP000015103 UP000002358 UP000078540 UP000008237 UP000192223 UP000008792 UP000053105 UP000053097 UP000005205 UP000007755 UP000015102 UP000183832 UP000027135 UP000242457 UP000008820 UP000076502 UP000053825 UP000030765 UP000001292 UP000000803 UP000092462 UP000075880 UP000001070 UP000092553 UP000008711 UP000007798 UP000000304 UP000075884 UP000002282 UP000092461 UP000268350 UP000009192 UP000001819 UP000192221 UP000007801 UP000075886 UP000008744 UP000005203 UP000075903 UP000007062 UP000075900 UP000075881 UP000076407 UP000092444 UP000092460 UP000075840 UP000092443 UP000030742 UP000075902 UP000095301 UP000001555
Pfam
PF05502 Dynactin_p62
ProteinModelPortal
H9IYP5
A0A2A4IV45
A0A3S2NAG8
A0A0N1IAF4
A0A194QPN2
A0A212EUT8
+ More
A0A2H1V6E3 A0A2J7PY88 A0A1Y1JZ50 D6X4D7 A0A3R7M8D9 A0A0P4VW45 T1IG42 A0A069DZ40 K7J4L4 A0A151I3X3 E2BDW5 A0A0C9R983 A0A0V0GAT3 A0A1B6JFK8 A0A1B6DNW3 A0A0P4W2P9 A0A224XBS8 A0A1W4WQT5 A0A1B6FV03 B4LJG9 A0A0N0BC75 A0A026WAB7 A0A158NYM2 E9JDH4 F4WXY9 T1GM44 A0A0K8TMS8 A0A1J1HKN3 A0A067QMH3 A0A1Q3G1N4 J7G8Y8 A0A1Q3G1Q4 A0A1Q3G1R1 Q17AJ7 A0A1Q3G1V0 A0A0A9X5U0 A0A1Q3G1K6 A0A0A1XLE9 A0A154NY09 A0A0L7RH23 W8CCU5 A0A0K8TWA1 A0A084VC99 B4HR53 Q7K130 A0A1B0GN13 A0A034W8J4 A0A182IWX8 B4J7Z7 A0A0M4EHH9 B3N9V6 B4N653 A0A0K8RIH6 V5HDN0 B4QEQ8 A0A182NED6 B4P1T3 A0A1B0CAC1 A0A0K8VVY5 A0A3B0J2Z4 B4KPR2 Q290Q3 A0A1B0CK06 A0A1W4WB48 B3MHS5 A0A182QQ57 B4GBE3 Q17AJ6 A0A1Z5KWK4 A0A088ADL4 A0A182URW3 Q7PNK5 A0A182RKC8 A0A182JXB4 A0A182XNK4 A0A1L8E5D8 A0A1B0GGF6 A0A1B0AT09 A0A182HMV7 A0A224YXK6 A0A131YPU0 A0A1A9YGV5 A0A131XNP4 U4TYN6 A0A182TUX7 A0A2M4AME9 A0A1I8N7K1 B7PEE3 T1JF52 D3TQ10
A0A2H1V6E3 A0A2J7PY88 A0A1Y1JZ50 D6X4D7 A0A3R7M8D9 A0A0P4VW45 T1IG42 A0A069DZ40 K7J4L4 A0A151I3X3 E2BDW5 A0A0C9R983 A0A0V0GAT3 A0A1B6JFK8 A0A1B6DNW3 A0A0P4W2P9 A0A224XBS8 A0A1W4WQT5 A0A1B6FV03 B4LJG9 A0A0N0BC75 A0A026WAB7 A0A158NYM2 E9JDH4 F4WXY9 T1GM44 A0A0K8TMS8 A0A1J1HKN3 A0A067QMH3 A0A1Q3G1N4 J7G8Y8 A0A1Q3G1Q4 A0A1Q3G1R1 Q17AJ7 A0A1Q3G1V0 A0A0A9X5U0 A0A1Q3G1K6 A0A0A1XLE9 A0A154NY09 A0A0L7RH23 W8CCU5 A0A0K8TWA1 A0A084VC99 B4HR53 Q7K130 A0A1B0GN13 A0A034W8J4 A0A182IWX8 B4J7Z7 A0A0M4EHH9 B3N9V6 B4N653 A0A0K8RIH6 V5HDN0 B4QEQ8 A0A182NED6 B4P1T3 A0A1B0CAC1 A0A0K8VVY5 A0A3B0J2Z4 B4KPR2 Q290Q3 A0A1B0CK06 A0A1W4WB48 B3MHS5 A0A182QQ57 B4GBE3 Q17AJ6 A0A1Z5KWK4 A0A088ADL4 A0A182URW3 Q7PNK5 A0A182RKC8 A0A182JXB4 A0A182XNK4 A0A1L8E5D8 A0A1B0GGF6 A0A1B0AT09 A0A182HMV7 A0A224YXK6 A0A131YPU0 A0A1A9YGV5 A0A131XNP4 U4TYN6 A0A182TUX7 A0A2M4AME9 A0A1I8N7K1 B7PEE3 T1JF52 D3TQ10
Ontologies
GO
PANTHER
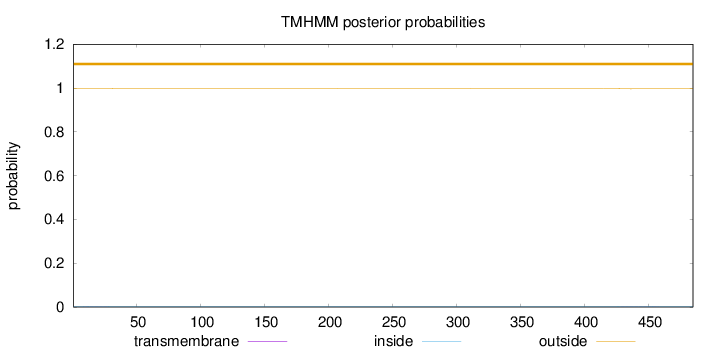
Topology
Length:
485
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0326400000000001
Exp number, first 60 AAs:
0.01285
Total prob of N-in:
0.00348
outside
1 - 485
Population Genetic Test Statistics
Pi
262.33143
Theta
193.392454
Tajima's D
0.887563
CLR
0
CSRT
0.635268236588171
Interpretation
Uncertain
