Gene
KWMTBOMO05358
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC105383094_[Plutella_xylostella]
Location in the cell
Extracellular Reliability : 1.505
Sequence
CDS
ATGTGTATCGGAGTCGATGGTGTGACTAGATTGACCAATGTAACACCACTACAATTTGAAGACAAATACGCATACAGCGCCACGCCGGTTATTGTCACTGATGCCACCGAAAAATGGCCAGCGATGAAGGTGTTCAACTTTAATTTCTTCTCGCGATTCTACAGCAATAATAACTTAGGTAGACAGACCAACGATTGTTTTTATTTCGCCTACAAATCCGGTCTGAATTCCCTGCAAGAAGTCTTCAATATGGATGAGAGACGCGCCAATTTATCCGGAGAACCGTGGTACGTAGGCTGGAGTACCTGCTTCGATGAGGAGACCAAGAAACTAAGATCGTATTACAATCGGCCGTATTTTCTCCCGCGGACGGCTGAGAGCGACACGGTGGACTGGATTTTTATGGGCGGACCCGGACAAGGGGCGCATATGCATGTGGACTCTGTAAGACACATGTCCTGGCAAGCGCAGATCCGCGGTCGCAAGCAATGGGAGCTGGCGCCCCCTCCTGAGTGCCTCTACCATTGCACCTGGATTCAATTCACAGTTTCTCCTGGTGAAATATTGGTCGTGAACACGAACCGTTGGTATCACAGGACTACCGTTCTCCCGGGAGACCTCAGCATCACGATCGGAGCTGAATATGATTAG
Protein
MCIGVDGVTRLTNVTPLQFEDKYAYSATPVIVTDATEKWPAMKVFNFNFFSRFYSNNNLGRQTNDCFYFAYKSGLNSLQEVFNMDERRANLSGEPWYVGWSTCFDEETKKLRSYYNRPYFLPRTAESDTVDWIFMGGPGQGAHMHVDSVRHMSWQAQIRGRKQWELAPPPECLYHCTWIQFTVSPGEILVVNTNRWYHRTTVLPGDLSITIGAEYD
Summary
Uniprot
A0A2A4IUH8
A0A2H1WF69
A0A3S2NQE9
A0A212EUU1
A0A0L7L7M6
A0A194QIR0
+ More
D6W8D8 A0A1B0GMQ6 A0A336MW11 A0A1B6GX48 A0A336MQ43 A0A1B0EZ43 Q16WD8 E0VB63 A0A1S4FLS1 A0A3R7P4T4 A0A182GLT2 A0A182RU61 A0A154PR71 A0A369SKQ9 B3RVH5 K7INM9 N6TST2 A0A182IED2 A0A182PJR8 A0A182X1F1 W5JA17 A0A182L2N7 Q7Q387 A0A182F1B2 A0A182UU22 A0A182JZ62 K1PAU1 A0A182T844 A0A182W7M1 A0A182MN78 A0A146M010 A0A0A9YN46 A0A1S4GWW8 A0A1B6DC48 B3LVQ8 B4K7M4 A0A1B6JQB9 A7S2W6 E2BJQ3 B4N9U4 A0A0L0BW64 A0A026X3R8 A0A2B4RLK8 A0A0L7QU52 A0A3B0JE55 A0A182IS25 A0A1I8NH41 Q29A84 A0A2G8KL70 A0A1I8NWL0 B4LY41 A0A1W4UT95 R7V5I5 A0A2B4SV86 E2AHM7 A0A0M8ZXS8 A0A3M6UCX3 A8JR34 D9PTR3 B4PVB6 B4JTV3 K1R3C7 B4K0P6 F4W6U1 V4ATW7 A0A195CWR2 A0A158NSB3 A0A151HZE3 A0A195FVG4 A0A151JPZ1 T1JIM1 A0A1S3HNH6 R7TA74 A0A151XK35 B7P5D7 A0A0P6AW36 A0A226EXM7 A0A1A9UEL8 V4AI56 A0A0L8FKB3 A0A1B0A7L7 A0A1A9X628 A0A2B4RL96 A0A1W4V6L8 A0A3M6UMQ9 W4Z679 A0A1B0C0M2 A0A267GDR7 E9GFY0 A0A0M3QXQ7 A0A210PX07 T1GE84 A0A1S3J463
D6W8D8 A0A1B0GMQ6 A0A336MW11 A0A1B6GX48 A0A336MQ43 A0A1B0EZ43 Q16WD8 E0VB63 A0A1S4FLS1 A0A3R7P4T4 A0A182GLT2 A0A182RU61 A0A154PR71 A0A369SKQ9 B3RVH5 K7INM9 N6TST2 A0A182IED2 A0A182PJR8 A0A182X1F1 W5JA17 A0A182L2N7 Q7Q387 A0A182F1B2 A0A182UU22 A0A182JZ62 K1PAU1 A0A182T844 A0A182W7M1 A0A182MN78 A0A146M010 A0A0A9YN46 A0A1S4GWW8 A0A1B6DC48 B3LVQ8 B4K7M4 A0A1B6JQB9 A7S2W6 E2BJQ3 B4N9U4 A0A0L0BW64 A0A026X3R8 A0A2B4RLK8 A0A0L7QU52 A0A3B0JE55 A0A182IS25 A0A1I8NH41 Q29A84 A0A2G8KL70 A0A1I8NWL0 B4LY41 A0A1W4UT95 R7V5I5 A0A2B4SV86 E2AHM7 A0A0M8ZXS8 A0A3M6UCX3 A8JR34 D9PTR3 B4PVB6 B4JTV3 K1R3C7 B4K0P6 F4W6U1 V4ATW7 A0A195CWR2 A0A158NSB3 A0A151HZE3 A0A195FVG4 A0A151JPZ1 T1JIM1 A0A1S3HNH6 R7TA74 A0A151XK35 B7P5D7 A0A0P6AW36 A0A226EXM7 A0A1A9UEL8 V4AI56 A0A0L8FKB3 A0A1B0A7L7 A0A1A9X628 A0A2B4RL96 A0A1W4V6L8 A0A3M6UMQ9 W4Z679 A0A1B0C0M2 A0A267GDR7 E9GFY0 A0A0M3QXQ7 A0A210PX07 T1GE84 A0A1S3J463
Pubmed
22118469
26227816
26354079
18362917
19820115
17510324
+ More
20566863 26483478 30042472 18719581 20075255 23537049 20920257 23761445 20966253 12364791 22992520 26823975 25401762 17994087 17615350 20798317 26108605 24508170 30249741 25315136 15632085 29023486 23254933 30382153 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 21719571 21347285 21292972 28812685 26383154
20566863 26483478 30042472 18719581 20075255 23537049 20920257 23761445 20966253 12364791 22992520 26823975 25401762 17994087 17615350 20798317 26108605 24508170 30249741 25315136 15632085 29023486 23254933 30382153 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 21719571 21347285 21292972 28812685 26383154
EMBL
NWSH01007004
PCG63156.1
ODYU01008280
SOQ51728.1
RSAL01000148
RVE45835.1
+ More
AGBW02012297 OWR45259.1 JTDY01002394 KOB71502.1 KQ458714 KPJ05443.1 KQ971307 EFA10937.1 AJVK01012234 UFQT01002933 SSX34310.1 GECZ01002789 JAS66980.1 UFQT01001407 SSX30417.1 AJWK01021849 CH477570 EAT38879.1 AAZO01000622 DS235022 EEB10619.1 QCYY01004272 ROT61238.1 JXUM01072955 JXUM01072956 JXUM01072957 KQ562750 KXJ75213.1 KQ435050 KZC14247.1 NOWV01000005 RDD47054.1 DS985244 EDV25502.1 APGK01020340 KB740193 KB631669 ENN81093.1 ERL85207.1 APCN01005201 ADMH02001723 ETN61302.1 AAAB01008964 EAA12909.2 JH823223 EKC18538.1 AXCM01006212 GDHC01005451 JAQ13178.1 GBHO01011081 GBHO01011080 JAG32523.1 JAG32524.1 GEDC01021008 GEDC01014062 JAS16290.1 JAS23236.1 CH902617 EDV43682.2 CH933806 EDW15368.2 GECU01006262 JAT01445.1 DS469570 EDO42000.1 GL448604 EFN84111.1 CH964232 EDW80659.2 JRES01001246 KNC24280.1 KK107019 QOIP01000007 EZA62728.1 RLU20148.1 LSMT01000495 PFX17132.1 KQ414735 KOC62150.1 OUUW01000005 SPP80654.1 CM000070 EAL27466.2 MRZV01000507 PIK48698.1 CH940650 EDW67929.1 AMQN01005862 KB297016 ELU11050.1 LSMT01000015 PFX33089.1 GL439566 EFN67061.1 KQ435800 KOX73285.1 RCHS01001758 RMX51547.1 AE014297 ABW08695.1 BT125070 ADK93998.1 CM000160 EDW95785.2 CH916374 EDV91532.1 JH816435 EKC35645.1 CH916484 EDV95100.1 GL887726 EGI70103.1 KB201262 ESO98350.1 KQ977171 KYN05118.1 ADTU01024765 ADTU01024766 KQ976699 KYM77411.1 KQ981241 KYN44287.1 KQ978724 KYN29027.1 AFFK01014481 AMQN01003055 KB310836 ELT90614.1 KQ982045 KYQ60764.1 ABJB010214470 DS640379 EEC01809.1 GDIP01023502 JAM80213.1 LNIX01000001 OXA61581.1 KB199905 ESP03764.1 KQ429904 KOF64958.1 PFX17127.1 RCHS01001150 RMX54946.1 AAGJ04168288 AAGJ04168289 AAGJ04168290 AAGJ04168291 JXJN01023690 NIVC01000387 PAA84136.1 GL732542 EFX81717.1 CP012526 ALC46289.1 NEDP02005430 OWF41020.1 CAQQ02027893
AGBW02012297 OWR45259.1 JTDY01002394 KOB71502.1 KQ458714 KPJ05443.1 KQ971307 EFA10937.1 AJVK01012234 UFQT01002933 SSX34310.1 GECZ01002789 JAS66980.1 UFQT01001407 SSX30417.1 AJWK01021849 CH477570 EAT38879.1 AAZO01000622 DS235022 EEB10619.1 QCYY01004272 ROT61238.1 JXUM01072955 JXUM01072956 JXUM01072957 KQ562750 KXJ75213.1 KQ435050 KZC14247.1 NOWV01000005 RDD47054.1 DS985244 EDV25502.1 APGK01020340 KB740193 KB631669 ENN81093.1 ERL85207.1 APCN01005201 ADMH02001723 ETN61302.1 AAAB01008964 EAA12909.2 JH823223 EKC18538.1 AXCM01006212 GDHC01005451 JAQ13178.1 GBHO01011081 GBHO01011080 JAG32523.1 JAG32524.1 GEDC01021008 GEDC01014062 JAS16290.1 JAS23236.1 CH902617 EDV43682.2 CH933806 EDW15368.2 GECU01006262 JAT01445.1 DS469570 EDO42000.1 GL448604 EFN84111.1 CH964232 EDW80659.2 JRES01001246 KNC24280.1 KK107019 QOIP01000007 EZA62728.1 RLU20148.1 LSMT01000495 PFX17132.1 KQ414735 KOC62150.1 OUUW01000005 SPP80654.1 CM000070 EAL27466.2 MRZV01000507 PIK48698.1 CH940650 EDW67929.1 AMQN01005862 KB297016 ELU11050.1 LSMT01000015 PFX33089.1 GL439566 EFN67061.1 KQ435800 KOX73285.1 RCHS01001758 RMX51547.1 AE014297 ABW08695.1 BT125070 ADK93998.1 CM000160 EDW95785.2 CH916374 EDV91532.1 JH816435 EKC35645.1 CH916484 EDV95100.1 GL887726 EGI70103.1 KB201262 ESO98350.1 KQ977171 KYN05118.1 ADTU01024765 ADTU01024766 KQ976699 KYM77411.1 KQ981241 KYN44287.1 KQ978724 KYN29027.1 AFFK01014481 AMQN01003055 KB310836 ELT90614.1 KQ982045 KYQ60764.1 ABJB010214470 DS640379 EEC01809.1 GDIP01023502 JAM80213.1 LNIX01000001 OXA61581.1 KB199905 ESP03764.1 KQ429904 KOF64958.1 PFX17127.1 RCHS01001150 RMX54946.1 AAGJ04168288 AAGJ04168289 AAGJ04168290 AAGJ04168291 JXJN01023690 NIVC01000387 PAA84136.1 GL732542 EFX81717.1 CP012526 ALC46289.1 NEDP02005430 OWF41020.1 CAQQ02027893
Proteomes
UP000218220
UP000283053
UP000007151
UP000037510
UP000053268
UP000007266
+ More
UP000092462 UP000092461 UP000008820 UP000009046 UP000283509 UP000069940 UP000249989 UP000075900 UP000076502 UP000253843 UP000009022 UP000002358 UP000019118 UP000030742 UP000075840 UP000075885 UP000076407 UP000000673 UP000075882 UP000007062 UP000069272 UP000075903 UP000075881 UP000005408 UP000075901 UP000075920 UP000075883 UP000007801 UP000009192 UP000001593 UP000008237 UP000007798 UP000037069 UP000053097 UP000279307 UP000225706 UP000053825 UP000268350 UP000075880 UP000095301 UP000001819 UP000230750 UP000095300 UP000008792 UP000192221 UP000014760 UP000000311 UP000053105 UP000275408 UP000000803 UP000002282 UP000001070 UP000007755 UP000030746 UP000078542 UP000005205 UP000078540 UP000078541 UP000078492 UP000085678 UP000075809 UP000001555 UP000198287 UP000078200 UP000053454 UP000092445 UP000092443 UP000007110 UP000092460 UP000215902 UP000000305 UP000092553 UP000242188 UP000015102
UP000092462 UP000092461 UP000008820 UP000009046 UP000283509 UP000069940 UP000249989 UP000075900 UP000076502 UP000253843 UP000009022 UP000002358 UP000019118 UP000030742 UP000075840 UP000075885 UP000076407 UP000000673 UP000075882 UP000007062 UP000069272 UP000075903 UP000075881 UP000005408 UP000075901 UP000075920 UP000075883 UP000007801 UP000009192 UP000001593 UP000008237 UP000007798 UP000037069 UP000053097 UP000279307 UP000225706 UP000053825 UP000268350 UP000075880 UP000095301 UP000001819 UP000230750 UP000095300 UP000008792 UP000192221 UP000014760 UP000000311 UP000053105 UP000275408 UP000000803 UP000002282 UP000001070 UP000007755 UP000030746 UP000078542 UP000005205 UP000078540 UP000078541 UP000078492 UP000085678 UP000075809 UP000001555 UP000198287 UP000078200 UP000053454 UP000092445 UP000092443 UP000007110 UP000092460 UP000215902 UP000000305 UP000092553 UP000242188 UP000015102
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IUH8
A0A2H1WF69
A0A3S2NQE9
A0A212EUU1
A0A0L7L7M6
A0A194QIR0
+ More
D6W8D8 A0A1B0GMQ6 A0A336MW11 A0A1B6GX48 A0A336MQ43 A0A1B0EZ43 Q16WD8 E0VB63 A0A1S4FLS1 A0A3R7P4T4 A0A182GLT2 A0A182RU61 A0A154PR71 A0A369SKQ9 B3RVH5 K7INM9 N6TST2 A0A182IED2 A0A182PJR8 A0A182X1F1 W5JA17 A0A182L2N7 Q7Q387 A0A182F1B2 A0A182UU22 A0A182JZ62 K1PAU1 A0A182T844 A0A182W7M1 A0A182MN78 A0A146M010 A0A0A9YN46 A0A1S4GWW8 A0A1B6DC48 B3LVQ8 B4K7M4 A0A1B6JQB9 A7S2W6 E2BJQ3 B4N9U4 A0A0L0BW64 A0A026X3R8 A0A2B4RLK8 A0A0L7QU52 A0A3B0JE55 A0A182IS25 A0A1I8NH41 Q29A84 A0A2G8KL70 A0A1I8NWL0 B4LY41 A0A1W4UT95 R7V5I5 A0A2B4SV86 E2AHM7 A0A0M8ZXS8 A0A3M6UCX3 A8JR34 D9PTR3 B4PVB6 B4JTV3 K1R3C7 B4K0P6 F4W6U1 V4ATW7 A0A195CWR2 A0A158NSB3 A0A151HZE3 A0A195FVG4 A0A151JPZ1 T1JIM1 A0A1S3HNH6 R7TA74 A0A151XK35 B7P5D7 A0A0P6AW36 A0A226EXM7 A0A1A9UEL8 V4AI56 A0A0L8FKB3 A0A1B0A7L7 A0A1A9X628 A0A2B4RL96 A0A1W4V6L8 A0A3M6UMQ9 W4Z679 A0A1B0C0M2 A0A267GDR7 E9GFY0 A0A0M3QXQ7 A0A210PX07 T1GE84 A0A1S3J463
D6W8D8 A0A1B0GMQ6 A0A336MW11 A0A1B6GX48 A0A336MQ43 A0A1B0EZ43 Q16WD8 E0VB63 A0A1S4FLS1 A0A3R7P4T4 A0A182GLT2 A0A182RU61 A0A154PR71 A0A369SKQ9 B3RVH5 K7INM9 N6TST2 A0A182IED2 A0A182PJR8 A0A182X1F1 W5JA17 A0A182L2N7 Q7Q387 A0A182F1B2 A0A182UU22 A0A182JZ62 K1PAU1 A0A182T844 A0A182W7M1 A0A182MN78 A0A146M010 A0A0A9YN46 A0A1S4GWW8 A0A1B6DC48 B3LVQ8 B4K7M4 A0A1B6JQB9 A7S2W6 E2BJQ3 B4N9U4 A0A0L0BW64 A0A026X3R8 A0A2B4RLK8 A0A0L7QU52 A0A3B0JE55 A0A182IS25 A0A1I8NH41 Q29A84 A0A2G8KL70 A0A1I8NWL0 B4LY41 A0A1W4UT95 R7V5I5 A0A2B4SV86 E2AHM7 A0A0M8ZXS8 A0A3M6UCX3 A8JR34 D9PTR3 B4PVB6 B4JTV3 K1R3C7 B4K0P6 F4W6U1 V4ATW7 A0A195CWR2 A0A158NSB3 A0A151HZE3 A0A195FVG4 A0A151JPZ1 T1JIM1 A0A1S3HNH6 R7TA74 A0A151XK35 B7P5D7 A0A0P6AW36 A0A226EXM7 A0A1A9UEL8 V4AI56 A0A0L8FKB3 A0A1B0A7L7 A0A1A9X628 A0A2B4RL96 A0A1W4V6L8 A0A3M6UMQ9 W4Z679 A0A1B0C0M2 A0A267GDR7 E9GFY0 A0A0M3QXQ7 A0A210PX07 T1GE84 A0A1S3J463
PDB
6FQC
E-value=0.0353076,
Score=83
Ontologies
KEGG
GO
PANTHER
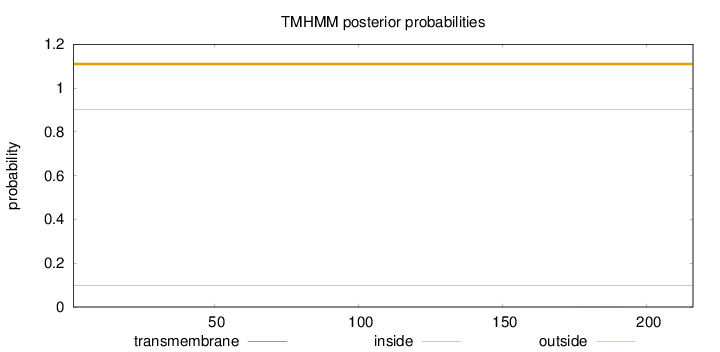
Topology
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00735
Exp number, first 60 AAs:
0.00116
Total prob of N-in:
0.09919
outside
1 - 216
Population Genetic Test Statistics
Pi
165.064568
Theta
130.716647
Tajima's D
-0.35186
CLR
0
CSRT
0.27863606819659
Interpretation
Uncertain
