Gene
KWMTBOMO05357 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002539
Annotation
PREDICTED:_V-type_proton_ATPase_116_kDa_subunit_a_isoform_1_[Bombyx_mori]
Full name
V-type proton ATPase subunit a
Location in the cell
PlasmaMembrane Reliability : 4.661
Sequence
CDS
ATGGGGTCTTTGTTTCGGAGTGAGGAGATGACACTATGTCAGCTTTTCCTGCAAAGTGAAGCTGCTTATGCCTGCGTGTCCGAGCTCGGGGAGTTGGGGCTAGTTCAGTTCCGAGATTTGAATCCTGACGTAAATGCCTTCCAACGTAAGTTCGTCAATGAAGTACGTCGCTGCGATGAGATGGAACGTAAGCTCCGCTACCTGGAGAAGGAGATCAGACGTGACGGCATCCCCATGCTGGAGATCCCCGGAGAGTGTCCCGAAGCGCCTCAACCCAGGGAGATGATCGATCTTGAAGCGACCTTCGAAAAGCTTGAGAATGAGCTGCGAGAGGTTAACCAGAACGCTGAGGCTCTTAAGAGGAACTACCTCGAGTTAACAGAATTAAAACACATTTTGAGGAAGACTCAAGTGTTCTTCGATGAGATGGCGGATCCGTCGAGGGAGGAGGAACAAGTCACCCTACTGGGGGAGGAGGGCCTCATGGCGGGAGGGCAAGCGCTCAAGCTGGGTTTCGTCGCGGGCGTGATTCTGAGGGAGAGAATACCGGCGTTCGAGCGCATGTTGTGGCGGGCCTGCAGGGGAAACGTGTTCCTGAGGCAGGCTGAGATCGACACGCCCCTGGAAGATCCATCATCATCGGATCAGGTTTACAAATCGGTGTTCATCATATTCTTCCAAGGCGACCAGCTGAAGACGCGCGTCAAGAAAATATGCGAAGGTTTCAGGGCGACGCTGTATCCGTGCCCGGAGTCGCCGGCCGACCGCCGGGAGATGGCCATGGGCGTCATGACCAGGATCGAGGATCTGAACACCGTCTTAGGTCAGACCCAGGACCATCGTCACCGCGTGCTGGTCGCCGCCGCCAAGAACATTAAGAACTGGTTCGTGAAAGTGCGCAAGATCAAGGCCATCTACCACACGTTGAATCTGTTCAATCTGGACGTGACCCAGAAGTGCCTCATCGCCGAGTGCTGGGTGCCCGCGCTCGACATGGAGACTATCCAACTGGCTCTGAGGAGAGGAACGGAACGCAGTGGCAGCTCGGTTCCCCCGATCCTCAACCGGATGGAGACTATAGAAGACCCGCCGACGTATAACAGGACCAATAAGTTCACTTCGGCCTTCCAGCACCTCATCTACGCGTACGGCGTGGCCACGTACCGCGAGGTCAACCCGGCACCCTACACGGTCATAACGTTCCCGTTCTTGTTCGCGGTGATGTTCGGCGACCTCGGCCACGGCGCCATCATGGCCGCCTTCGGGTTCTGGATGTGCTACAAGGAGAAGCCGCTGCAGGCCAAGAAAATCGACAGCGAGATCTGGAATATATTCTTCGGCGGTCGCTACATCATCCTGCTCATGGGCCTGTTCTCGATGTACACGGGCCTCATATACAACGACATATTCTCGAAGAGCTTGAACATATTCGGCTCGTCGTGGAGGAACAACTACGACGGCTCCACGCTGCAGAGCAACCAGCTGCTGCAGCTGAACCCCGACTCCAAAGACTACCTGCAGTACCCTTACCCGTTCGGCATCGACCCGGTCTGGCAGCTCGCAGAAGCCAACAAGATTATCTTCATGAACGGTTACAAGATGAAGATTTCTATCATAATAGGTGTCTTCCACATGTTGTTCGGTGTCTGTCTCTCGCTTTGGAATCATTTGTACTTCAAGCGTCGCATCAGCATCTACGTGGAGTTCATCCCCCAGATACTGTTCCTGAGCCTGCTGTTCTTCTACATGGTGCTGCTCATGTTCATCAAGTGGACCACGTACGGCGCCACGCCCGGACACTTCGGCAGCCAGGACCCCGAGATAGTGAAGACGAGCGCGTTCTGCGCCCCGTCCATCCTCATCACGTTCATCAACATGATGCTGTTCAAGCACGACAAGAACACGCGGCCGCAGTGCGACGACTTCATGTTCTCCGGACAGATGTTCATACAGAAGCTGTTCGTGATCGTGGCTCTGCTGTGCGTGCCCATCATGCTGTTCGGGAAGCCGTACTTCATCATGCGCGAGCAGAAGCAGCGCGCCAGACAAGGTCACCAGCCGGTAGAAGGTAACGCCGAGAACGGCACGGCGGGCGGCGCTCCCGTCCCGGCCTCGGGACATCACGACGAAGAGATCACCGAGGTCTTCATTCACCAGGCCATCCACACCATAGAGTTCGTGTTGGGCAGCGTGTCGCACACGGCGTCGTACCTGCGGCTGTGGGCGCTCTCCCTCGCGCACGCGCAGCTCGCTGAGGTCGCCTGGAACATGCTGCTCAGGAAGGGTCTGATGTCCAACGACTACCAGGGCGGGATCTTCCTGTACGTGGTGTTCGCGGGCTGGGCGGCCATCTCCGTCTCCATCCTCGTGCTCATGGAGGGGCTCTCGGCCTTCCTGCACACGCTGCGTCTGCACTGGGTCGAGTTCCAGAGCAAGTTCTACGGAGGAGAAGGCTACCTCTTCCAGCCCTTCTCGTTCGAGATCATTCTAGACTCGGCGGGTCAGGCAGAAGAATAA
Protein
MGSLFRSEEMTLCQLFLQSEAAYACVSELGELGLVQFRDLNPDVNAFQRKFVNEVRRCDEMERKLRYLEKEIRRDGIPMLEIPGECPEAPQPREMIDLEATFEKLENELREVNQNAEALKRNYLELTELKHILRKTQVFFDEMADPSREEEQVTLLGEEGLMAGGQALKLGFVAGVILRERIPAFERMLWRACRGNVFLRQAEIDTPLEDPSSSDQVYKSVFIIFFQGDQLKTRVKKICEGFRATLYPCPESPADRREMAMGVMTRIEDLNTVLGQTQDHRHRVLVAAAKNIKNWFVKVRKIKAIYHTLNLFNLDVTQKCLIAECWVPALDMETIQLALRRGTERSGSSVPPILNRMETIEDPPTYNRTNKFTSAFQHLIYAYGVATYREVNPAPYTVITFPFLFAVMFGDLGHGAIMAAFGFWMCYKEKPLQAKKIDSEIWNIFFGGRYIILLMGLFSMYTGLIYNDIFSKSLNIFGSSWRNNYDGSTLQSNQLLQLNPDSKDYLQYPYPFGIDPVWQLAEANKIIFMNGYKMKISIIIGVFHMLFGVCLSLWNHLYFKRRISIYVEFIPQILFLSLLFFYMVLLMFIKWTTYGATPGHFGSQDPEIVKTSAFCAPSILITFINMMLFKHDKNTRPQCDDFMFSGQMFIQKLFVIVALLCVPIMLFGKPYFIMREQKQRARQGHQPVEGNAENGTAGGAPVPASGHHDEEITEVFIHQAIHTIEFVLGSVSHTASYLRLWALSLAHAQLAEVAWNMLLRKGLMSNDYQGGIFLYVVFAGWAAISVSILVLMEGLSAFLHTLRLHWVEFQSKFYGGEGYLFQPFSFEIILDSAGQAEE
Summary
Description
Essential component of the vacuolar proton pump (V-ATPase), a multimeric enzyme that catalyzes the translocation of protons across the membranes. Required for assembly and activity of the V-ATPase.
Similarity
Belongs to the V-ATPase 116 kDa subunit family.
Uniprot
Q9U5M9
A0A2R8G1V7
A0A3S2L4Y8
A0A2H1WF76
A0A1E1WNG2
A0A2J7QFQ9
+ More
A0A2J7QFR4 E0V9Q8 A0A158NXX8 A0A151IVV4 A0A3L8DJ15 V9I760 A0A1E1WEP8 A0A084WR28 K7IYB2 A0A1Y1MAK7 A0A139WN05 K7IYB1 A0A2M4CS16 A0A1Y1MG49 K7IYB3 A0A2M3YZG4 A0A2M4BDD5 A0A182GWD2 A0A1Y0AWQ7 A0A182W305 F5HLG5 K7IYB0 A0A2J7QFT0 V5GRH0 A0A0A9ZE97 J9K3R8 A0A0C9S0C2 B0WEX4 A0A0K8T8E8 F5HLG3 A0A0A9ZEA0 A0A0C9S300 K7IYB4 A0A0K8T8E6 A0A0Q9X0Y9 A0A0Q9XCS9 A0A182F8F2 U4UFR4 A0A026X2Y9 A0A1I8N3G3 A0A1I8N3G6 A0A2M4A3S2 A0A1L8DF17 A0A2M3YZI3 A0A2M4BDR5 A0A182WVE5 A0A0M9A440 A0A182I925 A0A182V339 A0A0Q9X8P4 A0A0Q9X9R1 A0A1A9UQ47 A0A1I8NPJ1 A0A1I8NPJ9 A0A182MQW0 A0A182Q8Y9 W5JHE2 Q0IFY3 U5EY30 A0A182TZM2 W8AJG4 A0A1I8N3I1 A0A1I8N3G5 A0A182PQQ1 Q5TT36 A0A0A9Z8V9 A0A034VNS1 V9I8F4 A0A2A3E040 A0A0Q9X1I2 A0A1I8NPI7 A0A1Q3FFS3 A0A182KXZ5 A0A0A9Z8W6 A0A1I8NPJ3 A0A0Q9X2Q6 A0A1B6E585 A0A0A7DW87 A0A069DZK7 K7IYB5 A0A0K8T8T5 A0A0A1XBK4 A0A182R8D8 A0A1B6KBS1 A0A224X765 W8B5T4 A0A0P4VSP4 A0A1Y1MAZ1 A0A0Q9X0A0 A0A1B6H9J3 A0A0Q9X256 A0A0T6BAC8 A0A1B6FBA4 A0A2H8TQP4
A0A2J7QFR4 E0V9Q8 A0A158NXX8 A0A151IVV4 A0A3L8DJ15 V9I760 A0A1E1WEP8 A0A084WR28 K7IYB2 A0A1Y1MAK7 A0A139WN05 K7IYB1 A0A2M4CS16 A0A1Y1MG49 K7IYB3 A0A2M3YZG4 A0A2M4BDD5 A0A182GWD2 A0A1Y0AWQ7 A0A182W305 F5HLG5 K7IYB0 A0A2J7QFT0 V5GRH0 A0A0A9ZE97 J9K3R8 A0A0C9S0C2 B0WEX4 A0A0K8T8E8 F5HLG3 A0A0A9ZEA0 A0A0C9S300 K7IYB4 A0A0K8T8E6 A0A0Q9X0Y9 A0A0Q9XCS9 A0A182F8F2 U4UFR4 A0A026X2Y9 A0A1I8N3G3 A0A1I8N3G6 A0A2M4A3S2 A0A1L8DF17 A0A2M3YZI3 A0A2M4BDR5 A0A182WVE5 A0A0M9A440 A0A182I925 A0A182V339 A0A0Q9X8P4 A0A0Q9X9R1 A0A1A9UQ47 A0A1I8NPJ1 A0A1I8NPJ9 A0A182MQW0 A0A182Q8Y9 W5JHE2 Q0IFY3 U5EY30 A0A182TZM2 W8AJG4 A0A1I8N3I1 A0A1I8N3G5 A0A182PQQ1 Q5TT36 A0A0A9Z8V9 A0A034VNS1 V9I8F4 A0A2A3E040 A0A0Q9X1I2 A0A1I8NPI7 A0A1Q3FFS3 A0A182KXZ5 A0A0A9Z8W6 A0A1I8NPJ3 A0A0Q9X2Q6 A0A1B6E585 A0A0A7DW87 A0A069DZK7 K7IYB5 A0A0K8T8T5 A0A0A1XBK4 A0A182R8D8 A0A1B6KBS1 A0A224X765 W8B5T4 A0A0P4VSP4 A0A1Y1MAZ1 A0A0Q9X0A0 A0A1B6H9J3 A0A0Q9X256 A0A0T6BAC8 A0A1B6FBA4 A0A2H8TQP4
Pubmed
EMBL
AJ249390
CAB55500.1
KY921845
AVP73917.1
RSAL01000148
RVE45836.1
+ More
ODYU01008280 SOQ51729.1 GDQN01002673 JAT88381.1 NEVH01014852 PNF27426.1 PNF27427.1 DS234995 EEB10093.1 ADTU01003697 ADTU01003698 ADTU01003699 ADTU01003700 KQ980882 KYN11932.1 QOIP01000007 RLU20143.1 JR036121 AEY56933.1 GDQN01005584 JAT85470.1 ATLV01025906 ATLV01025907 KE525402 KFB52672.1 GEZM01036239 JAV82869.1 KQ971311 KYB29409.1 GGFL01003956 MBW68134.1 GEZM01036241 JAV82866.1 GGFM01000911 MBW21662.1 GGFJ01001919 MBW51060.1 JXUM01092931 JXUM01092932 JXUM01092933 JXUM01092934 KQ564021 KXJ72912.1 KY921838 ART29427.1 AAAB01008888 EGK97125.1 PNF27428.1 GALX01004254 JAB64212.1 GBHO01001936 GDHC01018959 JAG41668.1 JAP99669.1 ABLF02026125 GBYB01015260 JAG85027.1 DS231912 EDS25832.1 GBRD01003998 JAG61823.1 EGK97126.1 GBHO01001931 JAG41673.1 GBYB01015261 JAG85028.1 GBRD01003999 JAG61822.1 CH933806 KRG01557.1 KRG01555.1 KB632277 ERL91208.1 KK107019 EZA62635.1 GGFK01002088 MBW35409.1 GFDF01009160 JAV04924.1 GGFM01000938 MBW21689.1 GGFJ01002002 MBW51143.1 KQ435762 KOX75459.1 APCN01003164 KRG01556.1 KRG01554.1 AXCM01000016 AXCN02001184 ADMH02001252 ETN63486.1 CH477281 EAT44941.1 GANO01000629 JAB59242.1 GAMC01017875 GAMC01017871 JAB88680.1 EAL40624.3 GBHO01001932 GDHC01015774 JAG41672.1 JAQ02855.1 GAKP01013981 GAKP01013980 JAC44971.1 JR036120 AEY56932.1 KZ288510 PBC25118.1 CH964232 KRF99395.1 GFDL01008651 JAV26394.1 GBHO01001922 GDHC01012224 JAG41682.1 JAQ06405.1 KRF99393.1 GEDC01004206 JAS33092.1 KF986377 AIY24627.1 GBGD01000470 JAC88419.1 GBRD01004000 JAG61821.1 GBXI01005967 JAD08325.1 GEBQ01031086 JAT08891.1 GFTR01008131 JAW08295.1 GAMC01017874 JAB88681.1 GDKW01001098 JAI55497.1 GEZM01036243 JAV82864.1 KRG01558.1 GECU01036404 JAS71302.1 KRG01561.1 LJIG01002646 KRT84294.1 GECZ01022307 GECZ01013541 JAS47462.1 JAS56228.1 GFXV01004708 MBW16513.1
ODYU01008280 SOQ51729.1 GDQN01002673 JAT88381.1 NEVH01014852 PNF27426.1 PNF27427.1 DS234995 EEB10093.1 ADTU01003697 ADTU01003698 ADTU01003699 ADTU01003700 KQ980882 KYN11932.1 QOIP01000007 RLU20143.1 JR036121 AEY56933.1 GDQN01005584 JAT85470.1 ATLV01025906 ATLV01025907 KE525402 KFB52672.1 GEZM01036239 JAV82869.1 KQ971311 KYB29409.1 GGFL01003956 MBW68134.1 GEZM01036241 JAV82866.1 GGFM01000911 MBW21662.1 GGFJ01001919 MBW51060.1 JXUM01092931 JXUM01092932 JXUM01092933 JXUM01092934 KQ564021 KXJ72912.1 KY921838 ART29427.1 AAAB01008888 EGK97125.1 PNF27428.1 GALX01004254 JAB64212.1 GBHO01001936 GDHC01018959 JAG41668.1 JAP99669.1 ABLF02026125 GBYB01015260 JAG85027.1 DS231912 EDS25832.1 GBRD01003998 JAG61823.1 EGK97126.1 GBHO01001931 JAG41673.1 GBYB01015261 JAG85028.1 GBRD01003999 JAG61822.1 CH933806 KRG01557.1 KRG01555.1 KB632277 ERL91208.1 KK107019 EZA62635.1 GGFK01002088 MBW35409.1 GFDF01009160 JAV04924.1 GGFM01000938 MBW21689.1 GGFJ01002002 MBW51143.1 KQ435762 KOX75459.1 APCN01003164 KRG01556.1 KRG01554.1 AXCM01000016 AXCN02001184 ADMH02001252 ETN63486.1 CH477281 EAT44941.1 GANO01000629 JAB59242.1 GAMC01017875 GAMC01017871 JAB88680.1 EAL40624.3 GBHO01001932 GDHC01015774 JAG41672.1 JAQ02855.1 GAKP01013981 GAKP01013980 JAC44971.1 JR036120 AEY56932.1 KZ288510 PBC25118.1 CH964232 KRF99395.1 GFDL01008651 JAV26394.1 GBHO01001922 GDHC01012224 JAG41682.1 JAQ06405.1 KRF99393.1 GEDC01004206 JAS33092.1 KF986377 AIY24627.1 GBGD01000470 JAC88419.1 GBRD01004000 JAG61821.1 GBXI01005967 JAD08325.1 GEBQ01031086 JAT08891.1 GFTR01008131 JAW08295.1 GAMC01017874 JAB88681.1 GDKW01001098 JAI55497.1 GEZM01036243 JAV82864.1 KRG01558.1 GECU01036404 JAS71302.1 KRG01561.1 LJIG01002646 KRT84294.1 GECZ01022307 GECZ01013541 JAS47462.1 JAS56228.1 GFXV01004708 MBW16513.1
Proteomes
UP000283053
UP000235965
UP000009046
UP000005205
UP000078492
UP000279307
+ More
UP000030765 UP000002358 UP000007266 UP000069940 UP000249989 UP000075920 UP000007062 UP000007819 UP000002320 UP000009192 UP000069272 UP000030742 UP000053097 UP000095301 UP000053105 UP000075840 UP000075903 UP000078200 UP000095300 UP000075883 UP000075886 UP000000673 UP000008820 UP000075902 UP000075885 UP000242457 UP000007798 UP000075882 UP000075900
UP000030765 UP000002358 UP000007266 UP000069940 UP000249989 UP000075920 UP000007062 UP000007819 UP000002320 UP000009192 UP000069272 UP000030742 UP000053097 UP000095301 UP000053105 UP000075840 UP000075903 UP000078200 UP000095300 UP000075883 UP000075886 UP000000673 UP000008820 UP000075902 UP000075885 UP000242457 UP000007798 UP000075882 UP000075900
Pfam
PF01496 V_ATPase_I
ProteinModelPortal
Q9U5M9
A0A2R8G1V7
A0A3S2L4Y8
A0A2H1WF76
A0A1E1WNG2
A0A2J7QFQ9
+ More
A0A2J7QFR4 E0V9Q8 A0A158NXX8 A0A151IVV4 A0A3L8DJ15 V9I760 A0A1E1WEP8 A0A084WR28 K7IYB2 A0A1Y1MAK7 A0A139WN05 K7IYB1 A0A2M4CS16 A0A1Y1MG49 K7IYB3 A0A2M3YZG4 A0A2M4BDD5 A0A182GWD2 A0A1Y0AWQ7 A0A182W305 F5HLG5 K7IYB0 A0A2J7QFT0 V5GRH0 A0A0A9ZE97 J9K3R8 A0A0C9S0C2 B0WEX4 A0A0K8T8E8 F5HLG3 A0A0A9ZEA0 A0A0C9S300 K7IYB4 A0A0K8T8E6 A0A0Q9X0Y9 A0A0Q9XCS9 A0A182F8F2 U4UFR4 A0A026X2Y9 A0A1I8N3G3 A0A1I8N3G6 A0A2M4A3S2 A0A1L8DF17 A0A2M3YZI3 A0A2M4BDR5 A0A182WVE5 A0A0M9A440 A0A182I925 A0A182V339 A0A0Q9X8P4 A0A0Q9X9R1 A0A1A9UQ47 A0A1I8NPJ1 A0A1I8NPJ9 A0A182MQW0 A0A182Q8Y9 W5JHE2 Q0IFY3 U5EY30 A0A182TZM2 W8AJG4 A0A1I8N3I1 A0A1I8N3G5 A0A182PQQ1 Q5TT36 A0A0A9Z8V9 A0A034VNS1 V9I8F4 A0A2A3E040 A0A0Q9X1I2 A0A1I8NPI7 A0A1Q3FFS3 A0A182KXZ5 A0A0A9Z8W6 A0A1I8NPJ3 A0A0Q9X2Q6 A0A1B6E585 A0A0A7DW87 A0A069DZK7 K7IYB5 A0A0K8T8T5 A0A0A1XBK4 A0A182R8D8 A0A1B6KBS1 A0A224X765 W8B5T4 A0A0P4VSP4 A0A1Y1MAZ1 A0A0Q9X0A0 A0A1B6H9J3 A0A0Q9X256 A0A0T6BAC8 A0A1B6FBA4 A0A2H8TQP4
A0A2J7QFR4 E0V9Q8 A0A158NXX8 A0A151IVV4 A0A3L8DJ15 V9I760 A0A1E1WEP8 A0A084WR28 K7IYB2 A0A1Y1MAK7 A0A139WN05 K7IYB1 A0A2M4CS16 A0A1Y1MG49 K7IYB3 A0A2M3YZG4 A0A2M4BDD5 A0A182GWD2 A0A1Y0AWQ7 A0A182W305 F5HLG5 K7IYB0 A0A2J7QFT0 V5GRH0 A0A0A9ZE97 J9K3R8 A0A0C9S0C2 B0WEX4 A0A0K8T8E8 F5HLG3 A0A0A9ZEA0 A0A0C9S300 K7IYB4 A0A0K8T8E6 A0A0Q9X0Y9 A0A0Q9XCS9 A0A182F8F2 U4UFR4 A0A026X2Y9 A0A1I8N3G3 A0A1I8N3G6 A0A2M4A3S2 A0A1L8DF17 A0A2M3YZI3 A0A2M4BDR5 A0A182WVE5 A0A0M9A440 A0A182I925 A0A182V339 A0A0Q9X8P4 A0A0Q9X9R1 A0A1A9UQ47 A0A1I8NPJ1 A0A1I8NPJ9 A0A182MQW0 A0A182Q8Y9 W5JHE2 Q0IFY3 U5EY30 A0A182TZM2 W8AJG4 A0A1I8N3I1 A0A1I8N3G5 A0A182PQQ1 Q5TT36 A0A0A9Z8V9 A0A034VNS1 V9I8F4 A0A2A3E040 A0A0Q9X1I2 A0A1I8NPI7 A0A1Q3FFS3 A0A182KXZ5 A0A0A9Z8W6 A0A1I8NPJ3 A0A0Q9X2Q6 A0A1B6E585 A0A0A7DW87 A0A069DZK7 K7IYB5 A0A0K8T8T5 A0A0A1XBK4 A0A182R8D8 A0A1B6KBS1 A0A224X765 W8B5T4 A0A0P4VSP4 A0A1Y1MAZ1 A0A0Q9X0A0 A0A1B6H9J3 A0A0Q9X256 A0A0T6BAC8 A0A1B6FBA4 A0A2H8TQP4
PDB
6O7T
E-value=1.24199e-155,
Score=1413
Ontologies
PATHWAY
GO
PANTHER
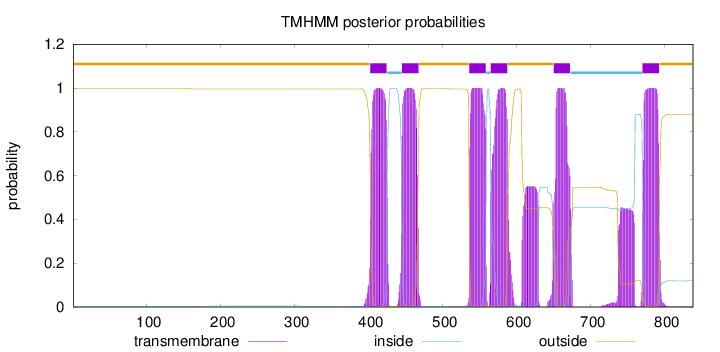
Topology
Length:
838
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
156.16653
Exp number, first 60 AAs:
0.00088
Total prob of N-in:
0.00353
outside
1 - 401
TMhelix
402 - 424
inside
425 - 444
TMhelix
445 - 467
outside
468 - 535
TMhelix
536 - 558
inside
559 - 564
TMhelix
565 - 587
outside
588 - 649
TMhelix
650 - 672
inside
673 - 769
TMhelix
770 - 792
outside
793 - 838
Population Genetic Test Statistics
Pi
199.094363
Theta
159.245694
Tajima's D
0.75394
CLR
0.561409
CSRT
0.587770611469427
Interpretation
Uncertain
