Gene
KWMTBOMO05356
Pre Gene Modal
BGIBMGA002380
Annotation
PREDICTED:_transmembrane_protein_161B_isoform_X1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.87
Sequence
CDS
ATGGCATTATTGGGGGCTCAGCTTGTAATAACTTTGATCATGGTATCAGTTATACAGAAATTGGGCAATTATTCGTTCGCGAAGTGGTTGCTGTGCTCTCAGGGCTTATACCGGTATTTGTATCCGGACAATAATGAGTTGAAGAATTTGGCTGGGGTTCCTAAAGAGAGATCTAAGGGGAAAAAAGGCGGTAAATATGAGAGTAACGGTAAACCGGAGACATTTCACGTGCCTAGAAGCTTGGAAATCCAATTGGAAACTGCACCGGTTACTGCGCTGGACGTGGTACATTTGAGATTCTACACAGAATATCTTTGGATTGTTGATTTTTCATTGTATACAGCCATAGTATACATTATGTCTGAGGTTTACACCTCAATATTTCCATTAAAAGATGAGTTCAATTTGAGCATGGTCTGGTGTCTACTCGTTATTTTGTTTTCAATTAAAATCTTGTTATCACTTACCAAACAATATTTCACAAGTGATGAATCAATCGGTGAACGATCAACATGCATTGTCGCATTCTGCGTTTTCCTCCTCATAGCCATGATGGTACTGATCGTAGATGAAAAAAACTTGGAAGTTGGTGTGGACCCTGCTTATGACAGCTTTTTTGAAAATGCTTCTAAGTTTCTCGATAGTCAAGGCTTGAGTTCTGTTGGTCCAGCATCCAAGCTTGTGATAAAGTTTGCTTTTGCCGTGTGGGCTGCGATCATTGGTACCATATTTACATTTCCGGGATTCAGAGTTTCCAGAATGCATTGGGACTCACTGAGGTACTACGGAGAAAACAAAATAAAGTATTTGTTGCTAAATTTTAATTTTGCTCTGCCGTTCATACTTGTACTGTTGTGGGTGCGTCCAATTACGAGGCACTACCTCACAGCTCGAGTATTCAGCGGGATGGAGAAACCTTTAATGACGTCGCAGGCCTTCGACACCCTGCGCATCGTGCTGGTGGTGTGCACGGCGGCGCTGCGCGTGGCGCTGATGCCGCGGCAGCTGCAGGCCTACCTGGACATTGCACAGCGGAGGCTCGACGTGCAGAAGAAAGAGGCCGGCAGGATAACCAATATCGATTTACAGAAAAAGGTCGCCTCGGTGTTCTACTACCTGTGCGTGGTGGCGCTGCAGTACATCTGCCCCGTGATCATGATGCTGTACTTGGCCCTGATGTACAAGACGCTGGGCGGCCACGCGTGGAGCAGCCTGCTGTACGAGGCGCCCCCCGCCGCGCCCCTCCCCGCGCCCCCCGCCGCACCCCCCGCCGCCCCCGCAGGCGCGGAGCAGTACCAGCTGGCCTGGGAAAACTTGAAGATGGTGTTCACGACGGAGGTGTGTCGCGGAGTGCTGGGCTTCGCGACGTGGTGGTGCAGCTTCGCGTGGTTCGTGACCACCGCGCTGGGAGCCGCCTACCAGAGTTACTTCAGGATCTCCTGA
Protein
MALLGAQLVITLIMVSVIQKLGNYSFAKWLLCSQGLYRYLYPDNNELKNLAGVPKERSKGKKGGKYESNGKPETFHVPRSLEIQLETAPVTALDVVHLRFYTEYLWIVDFSLYTAIVYIMSEVYTSIFPLKDEFNLSMVWCLLVILFSIKILLSLTKQYFTSDESIGERSTCIVAFCVFLLIAMMVLIVDEKNLEVGVDPAYDSFFENASKFLDSQGLSSVGPASKLVIKFAFAVWAAIIGTIFTFPGFRVSRMHWDSLRYYGENKIKYLLLNFNFALPFILVLLWVRPITRHYLTARVFSGMEKPLMTSQAFDTLRIVLVVCTAALRVALMPRQLQAYLDIAQRRLDVQKKEAGRITNIDLQKKVASVFYYLCVVALQYICPVIMMLYLALMYKTLGGHAWSSLLYEAPPAAPLPAPPAAPPAAPAGAEQYQLAWENLKMVFTTEVCRGVLGFATWWCSFAWFVTTALGAAYQSYFRIS
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
A0A2H1WF68
A0A2A4IZH5
A0A194QKJ9
A0A212EUW9
A0A2M4AK68
A0A2J7RAJ4
+ More
A0A1B6JMN0 A0A1B6M1Y1 A0A067RJW2 A0A139WAU9 A0A2M4BKH9 W5J5M8 Q17LS9 A0A1B6E2L2 A0A023F2J4 A0A1S4EY72 A0A182GLM8 A0A1Q3F8Y6 A0A0N0U7B9 A0A1L8D9L6 U5EX94 E9IZE5 A0A336MVM4 A0A0C9R1F6 F4WD61 A0A1B6FZ93 A0A336LYD7 A0A158P0V6 B0WHN3 K7IX96 A0A310SDA2 A0A2C9GR10 Q7PPG5 A0A088AF64 A0A2J7RAJ8 A0A1B0C954 A0A2A3EPH6 A0A1J1IEY0 A0A146L5R8 A0A151JXI8 A0A195AUL0 A0A151ITV3 A0A3B0JUT4 A0A0L0BUG9 A0A182LYP9 B4L174 A0A1Y1KRZ9 A0A151WVY7 A0A2J7RAJ2 A0A1A9XGF7 E2BE57 A0A1B0BZM8 B4GZP0 Q2LZ28 A0A1A9ZKP3 A0A1B0FKE4 A0A1A9UFU6 B4N5E3 A0A1I8PRS8 B4LC45 A0A1I8M4W5 A0A3B0JPR4 B4QPQ0 B3M3W5 B4PEM0 B4HDW4 A0A182P5G5 A0A0J7KAD4 A0A1W4W4P9 B3NGR7 E0VKA5 Q9VTF5 A0A3L8DFB1 A0A0M4EL65 A0A2M4BML7 A0A182K4N4 A0A182X2H7 A0A182RAL7 B4IXZ9 A0A1B0D1L1 A0A182JMV3 A0A182N1Z7 A0A224XNS7 A0A026WFQ9 A0A182WM09 A0A182YKR9 A0A0A1X3U7 A0A084WD10 A0A0K2U7A8 W8ASG1 A0A0K8VJL0 E2AYY5 J9JWR0 A0A0V0GCF6 A0A1D2MT25 N6UDC1 A0A0P5K1S4 A0A2H8TWI8 A0A164L9C3 A0A0P4W7N2
A0A1B6JMN0 A0A1B6M1Y1 A0A067RJW2 A0A139WAU9 A0A2M4BKH9 W5J5M8 Q17LS9 A0A1B6E2L2 A0A023F2J4 A0A1S4EY72 A0A182GLM8 A0A1Q3F8Y6 A0A0N0U7B9 A0A1L8D9L6 U5EX94 E9IZE5 A0A336MVM4 A0A0C9R1F6 F4WD61 A0A1B6FZ93 A0A336LYD7 A0A158P0V6 B0WHN3 K7IX96 A0A310SDA2 A0A2C9GR10 Q7PPG5 A0A088AF64 A0A2J7RAJ8 A0A1B0C954 A0A2A3EPH6 A0A1J1IEY0 A0A146L5R8 A0A151JXI8 A0A195AUL0 A0A151ITV3 A0A3B0JUT4 A0A0L0BUG9 A0A182LYP9 B4L174 A0A1Y1KRZ9 A0A151WVY7 A0A2J7RAJ2 A0A1A9XGF7 E2BE57 A0A1B0BZM8 B4GZP0 Q2LZ28 A0A1A9ZKP3 A0A1B0FKE4 A0A1A9UFU6 B4N5E3 A0A1I8PRS8 B4LC45 A0A1I8M4W5 A0A3B0JPR4 B4QPQ0 B3M3W5 B4PEM0 B4HDW4 A0A182P5G5 A0A0J7KAD4 A0A1W4W4P9 B3NGR7 E0VKA5 Q9VTF5 A0A3L8DFB1 A0A0M4EL65 A0A2M4BML7 A0A182K4N4 A0A182X2H7 A0A182RAL7 B4IXZ9 A0A1B0D1L1 A0A182JMV3 A0A182N1Z7 A0A224XNS7 A0A026WFQ9 A0A182WM09 A0A182YKR9 A0A0A1X3U7 A0A084WD10 A0A0K2U7A8 W8ASG1 A0A0K8VJL0 E2AYY5 J9JWR0 A0A0V0GCF6 A0A1D2MT25 N6UDC1 A0A0P5K1S4 A0A2H8TWI8 A0A164L9C3 A0A0P4W7N2
Pubmed
26354079
22118469
24845553
18362917
19820115
20920257
+ More
23761445 17510324 25474469 26483478 21282665 21719571 21347285 20075255 12364791 14747013 17210077 26823975 26108605 17994087 18057021 28004739 20798317 15632085 25315136 22936249 17550304 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 24508170 25244985 25830018 24438588 24495485 27289101 23537049
23761445 17510324 25474469 26483478 21282665 21719571 21347285 20075255 12364791 14747013 17210077 26823975 26108605 17994087 18057021 28004739 20798317 15632085 25315136 22936249 17550304 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 24508170 25244985 25830018 24438588 24495485 27289101 23537049
EMBL
ODYU01008280
SOQ51730.1
NWSH01004819
PCG64674.1
KQ458714
KPJ05445.1
+ More
AGBW02012297 OWR45261.1 GGFK01007826 MBW41147.1 NEVH01006567 PNF37856.1 GECU01007170 JAT00537.1 GEBQ01010069 JAT29908.1 KK852427 KDR24097.1 KQ971380 KYB25035.1 GGFJ01004391 MBW53532.1 ADMH02002171 ETN58074.1 CH477213 EAT47622.1 GEDC01005157 JAS32141.1 GBBI01003371 JAC15341.1 JXUM01072365 JXUM01072366 KQ562717 KXJ75260.1 GFDL01011033 JAV24012.1 KQ435706 KOX80271.1 GFDF01011080 JAV03004.1 GANO01000178 JAB59693.1 GL767121 EFZ14096.1 UFQS01001316 UFQT01001316 SSX10310.1 SSX29998.1 GBYB01001770 JAG71537.1 GL888084 EGI67869.1 GECZ01016290 GECZ01014308 JAS53479.1 JAS55461.1 UFQS01000224 UFQT01000224 SSX01641.1 SSX22021.1 ADTU01005844 DS231938 EDS27882.1 AAZX01000617 KQ767877 OAD53283.1 APCN01004639 AAAB01008948 EAA10461.5 PNF37857.1 AJWK01001916 KZ288199 PBC33638.1 CVRI01000048 CRK98821.1 GDHC01014805 JAQ03824.1 KQ981572 KYN39767.1 KQ976738 KYM75705.1 KQ980991 KYN10470.1 OUUW01000002 SPP77469.1 JRES01001310 KNC23676.1 AXCM01003267 CH933809 EDW19256.2 KRG06572.1 GEZM01075242 GEZM01075241 JAV64182.1 KQ982691 KYQ52099.1 PNF37855.1 GL447755 EFN86044.1 JXJN01023210 CH479199 EDW29467.1 CH379069 EAL29681.1 CCAG010018197 CH964101 EDW79582.1 CH940647 EDW70873.2 SPP77470.1 CM000363 CM002912 EDX10041.1 KMY98930.1 CH902618 EDV39299.1 CM000159 EDW94086.1 CH480815 EDW41054.1 LBMM01010760 KMQ87257.1 CH954178 EDV51374.1 DS235241 EEB13811.1 AE014296 AY118392 AAF50096.2 AAM48421.1 QOIP01000009 RLU18608.1 CP012525 ALC44135.1 GGFJ01005062 MBW54203.1 CH916366 EDV97542.1 AJVK01022111 GFTR01006204 JAW10222.1 KK107235 EZA54887.1 GBXI01008984 JAD05308.1 ATLV01022908 KE525337 KFB48104.1 HACA01016549 CDW33910.1 GAMC01017513 JAB89042.1 GDHF01013228 GDHF01006369 JAI39086.1 JAI45945.1 GL444028 EFN61355.1 ABLF02027828 GECL01000217 JAP05907.1 LJIJ01000569 ODM96166.1 APGK01039372 APGK01039373 KB740970 ENN76652.1 GDIQ01190953 JAK60772.1 GFXV01006474 MBW18279.1 LRGB01003163 KZS03909.1 GDRN01109103 GDRN01109102 JAI57129.1
AGBW02012297 OWR45261.1 GGFK01007826 MBW41147.1 NEVH01006567 PNF37856.1 GECU01007170 JAT00537.1 GEBQ01010069 JAT29908.1 KK852427 KDR24097.1 KQ971380 KYB25035.1 GGFJ01004391 MBW53532.1 ADMH02002171 ETN58074.1 CH477213 EAT47622.1 GEDC01005157 JAS32141.1 GBBI01003371 JAC15341.1 JXUM01072365 JXUM01072366 KQ562717 KXJ75260.1 GFDL01011033 JAV24012.1 KQ435706 KOX80271.1 GFDF01011080 JAV03004.1 GANO01000178 JAB59693.1 GL767121 EFZ14096.1 UFQS01001316 UFQT01001316 SSX10310.1 SSX29998.1 GBYB01001770 JAG71537.1 GL888084 EGI67869.1 GECZ01016290 GECZ01014308 JAS53479.1 JAS55461.1 UFQS01000224 UFQT01000224 SSX01641.1 SSX22021.1 ADTU01005844 DS231938 EDS27882.1 AAZX01000617 KQ767877 OAD53283.1 APCN01004639 AAAB01008948 EAA10461.5 PNF37857.1 AJWK01001916 KZ288199 PBC33638.1 CVRI01000048 CRK98821.1 GDHC01014805 JAQ03824.1 KQ981572 KYN39767.1 KQ976738 KYM75705.1 KQ980991 KYN10470.1 OUUW01000002 SPP77469.1 JRES01001310 KNC23676.1 AXCM01003267 CH933809 EDW19256.2 KRG06572.1 GEZM01075242 GEZM01075241 JAV64182.1 KQ982691 KYQ52099.1 PNF37855.1 GL447755 EFN86044.1 JXJN01023210 CH479199 EDW29467.1 CH379069 EAL29681.1 CCAG010018197 CH964101 EDW79582.1 CH940647 EDW70873.2 SPP77470.1 CM000363 CM002912 EDX10041.1 KMY98930.1 CH902618 EDV39299.1 CM000159 EDW94086.1 CH480815 EDW41054.1 LBMM01010760 KMQ87257.1 CH954178 EDV51374.1 DS235241 EEB13811.1 AE014296 AY118392 AAF50096.2 AAM48421.1 QOIP01000009 RLU18608.1 CP012525 ALC44135.1 GGFJ01005062 MBW54203.1 CH916366 EDV97542.1 AJVK01022111 GFTR01006204 JAW10222.1 KK107235 EZA54887.1 GBXI01008984 JAD05308.1 ATLV01022908 KE525337 KFB48104.1 HACA01016549 CDW33910.1 GAMC01017513 JAB89042.1 GDHF01013228 GDHF01006369 JAI39086.1 JAI45945.1 GL444028 EFN61355.1 ABLF02027828 GECL01000217 JAP05907.1 LJIJ01000569 ODM96166.1 APGK01039372 APGK01039373 KB740970 ENN76652.1 GDIQ01190953 JAK60772.1 GFXV01006474 MBW18279.1 LRGB01003163 KZS03909.1 GDRN01109103 GDRN01109102 JAI57129.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000235965
UP000027135
UP000007266
+ More
UP000000673 UP000008820 UP000069940 UP000249989 UP000053105 UP000007755 UP000005205 UP000002320 UP000002358 UP000075840 UP000007062 UP000005203 UP000092461 UP000242457 UP000183832 UP000078541 UP000078540 UP000078492 UP000268350 UP000037069 UP000075883 UP000009192 UP000075809 UP000092443 UP000008237 UP000092460 UP000008744 UP000001819 UP000092445 UP000092444 UP000078200 UP000007798 UP000095300 UP000008792 UP000095301 UP000000304 UP000007801 UP000002282 UP000001292 UP000075885 UP000036403 UP000192221 UP000008711 UP000009046 UP000000803 UP000279307 UP000092553 UP000075881 UP000076407 UP000075900 UP000001070 UP000092462 UP000075880 UP000075884 UP000053097 UP000075920 UP000076408 UP000030765 UP000000311 UP000007819 UP000094527 UP000019118 UP000076858
UP000000673 UP000008820 UP000069940 UP000249989 UP000053105 UP000007755 UP000005205 UP000002320 UP000002358 UP000075840 UP000007062 UP000005203 UP000092461 UP000242457 UP000183832 UP000078541 UP000078540 UP000078492 UP000268350 UP000037069 UP000075883 UP000009192 UP000075809 UP000092443 UP000008237 UP000092460 UP000008744 UP000001819 UP000092445 UP000092444 UP000078200 UP000007798 UP000095300 UP000008792 UP000095301 UP000000304 UP000007801 UP000002282 UP000001292 UP000075885 UP000036403 UP000192221 UP000008711 UP000009046 UP000000803 UP000279307 UP000092553 UP000075881 UP000076407 UP000075900 UP000001070 UP000092462 UP000075880 UP000075884 UP000053097 UP000075920 UP000076408 UP000030765 UP000000311 UP000007819 UP000094527 UP000019118 UP000076858
PRIDE
Pfam
Interpro
IPR019395
Transmembrane_161A/B
+ More
IPR029048 HSP70_C_sf
IPR018181 Heat_shock_70_CS
IPR029047 HSP70_peptide-bd_sf
IPR013126 Hsp_70_fam
IPR019137 Nck-associated_protein-1
IPR005998 Ribosomal_L7_euk
IPR023106 Ribosomal_L30_central_dom_arc
IPR016082 Ribosomal_L30_ferredoxin-like
IPR012988 Ribosomal_L30_N
IPR018038 Ribosomal_L30_CS
IPR036919 L30_ferredoxin-like_sf
IPR035808 Ribosomal_L7_euk_arc
IPR029048 HSP70_C_sf
IPR018181 Heat_shock_70_CS
IPR029047 HSP70_peptide-bd_sf
IPR013126 Hsp_70_fam
IPR019137 Nck-associated_protein-1
IPR005998 Ribosomal_L7_euk
IPR023106 Ribosomal_L30_central_dom_arc
IPR016082 Ribosomal_L30_ferredoxin-like
IPR012988 Ribosomal_L30_N
IPR018038 Ribosomal_L30_CS
IPR036919 L30_ferredoxin-like_sf
IPR035808 Ribosomal_L7_euk_arc
Gene 3D
ProteinModelPortal
A0A2H1WF68
A0A2A4IZH5
A0A194QKJ9
A0A212EUW9
A0A2M4AK68
A0A2J7RAJ4
+ More
A0A1B6JMN0 A0A1B6M1Y1 A0A067RJW2 A0A139WAU9 A0A2M4BKH9 W5J5M8 Q17LS9 A0A1B6E2L2 A0A023F2J4 A0A1S4EY72 A0A182GLM8 A0A1Q3F8Y6 A0A0N0U7B9 A0A1L8D9L6 U5EX94 E9IZE5 A0A336MVM4 A0A0C9R1F6 F4WD61 A0A1B6FZ93 A0A336LYD7 A0A158P0V6 B0WHN3 K7IX96 A0A310SDA2 A0A2C9GR10 Q7PPG5 A0A088AF64 A0A2J7RAJ8 A0A1B0C954 A0A2A3EPH6 A0A1J1IEY0 A0A146L5R8 A0A151JXI8 A0A195AUL0 A0A151ITV3 A0A3B0JUT4 A0A0L0BUG9 A0A182LYP9 B4L174 A0A1Y1KRZ9 A0A151WVY7 A0A2J7RAJ2 A0A1A9XGF7 E2BE57 A0A1B0BZM8 B4GZP0 Q2LZ28 A0A1A9ZKP3 A0A1B0FKE4 A0A1A9UFU6 B4N5E3 A0A1I8PRS8 B4LC45 A0A1I8M4W5 A0A3B0JPR4 B4QPQ0 B3M3W5 B4PEM0 B4HDW4 A0A182P5G5 A0A0J7KAD4 A0A1W4W4P9 B3NGR7 E0VKA5 Q9VTF5 A0A3L8DFB1 A0A0M4EL65 A0A2M4BML7 A0A182K4N4 A0A182X2H7 A0A182RAL7 B4IXZ9 A0A1B0D1L1 A0A182JMV3 A0A182N1Z7 A0A224XNS7 A0A026WFQ9 A0A182WM09 A0A182YKR9 A0A0A1X3U7 A0A084WD10 A0A0K2U7A8 W8ASG1 A0A0K8VJL0 E2AYY5 J9JWR0 A0A0V0GCF6 A0A1D2MT25 N6UDC1 A0A0P5K1S4 A0A2H8TWI8 A0A164L9C3 A0A0P4W7N2
A0A1B6JMN0 A0A1B6M1Y1 A0A067RJW2 A0A139WAU9 A0A2M4BKH9 W5J5M8 Q17LS9 A0A1B6E2L2 A0A023F2J4 A0A1S4EY72 A0A182GLM8 A0A1Q3F8Y6 A0A0N0U7B9 A0A1L8D9L6 U5EX94 E9IZE5 A0A336MVM4 A0A0C9R1F6 F4WD61 A0A1B6FZ93 A0A336LYD7 A0A158P0V6 B0WHN3 K7IX96 A0A310SDA2 A0A2C9GR10 Q7PPG5 A0A088AF64 A0A2J7RAJ8 A0A1B0C954 A0A2A3EPH6 A0A1J1IEY0 A0A146L5R8 A0A151JXI8 A0A195AUL0 A0A151ITV3 A0A3B0JUT4 A0A0L0BUG9 A0A182LYP9 B4L174 A0A1Y1KRZ9 A0A151WVY7 A0A2J7RAJ2 A0A1A9XGF7 E2BE57 A0A1B0BZM8 B4GZP0 Q2LZ28 A0A1A9ZKP3 A0A1B0FKE4 A0A1A9UFU6 B4N5E3 A0A1I8PRS8 B4LC45 A0A1I8M4W5 A0A3B0JPR4 B4QPQ0 B3M3W5 B4PEM0 B4HDW4 A0A182P5G5 A0A0J7KAD4 A0A1W4W4P9 B3NGR7 E0VKA5 Q9VTF5 A0A3L8DFB1 A0A0M4EL65 A0A2M4BML7 A0A182K4N4 A0A182X2H7 A0A182RAL7 B4IXZ9 A0A1B0D1L1 A0A182JMV3 A0A182N1Z7 A0A224XNS7 A0A026WFQ9 A0A182WM09 A0A182YKR9 A0A0A1X3U7 A0A084WD10 A0A0K2U7A8 W8ASG1 A0A0K8VJL0 E2AYY5 J9JWR0 A0A0V0GCF6 A0A1D2MT25 N6UDC1 A0A0P5K1S4 A0A2H8TWI8 A0A164L9C3 A0A0P4W7N2
Ontologies
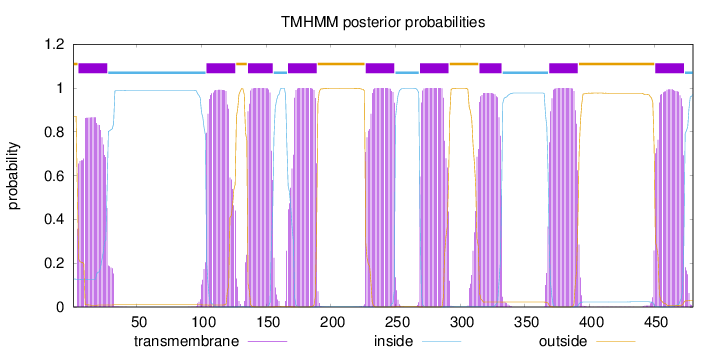
Topology
Length:
480
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
188.90274
Exp number, first 60 AAs:
19.0416
Total prob of N-in:
0.12930
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 103
TMhelix
104 - 126
outside
127 - 135
TMhelix
136 - 155
inside
156 - 166
TMhelix
167 - 189
outside
190 - 226
TMhelix
227 - 249
inside
250 - 268
TMhelix
269 - 291
outside
292 - 314
TMhelix
315 - 332
inside
333 - 368
TMhelix
369 - 391
outside
392 - 450
TMhelix
451 - 473
inside
474 - 480
Population Genetic Test Statistics
Pi
256.059167
Theta
161.964579
Tajima's D
2.003784
CLR
0.581844
CSRT
0.88355582220889
Interpretation
Uncertain
