Gene
KWMTBOMO05355 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002381
Annotation
heat_shock_cognate_protein_[Bombyx_mori]
Full name
Heat shock 70 kDa protein cognate 4
Location in the cell
Cytoplasmic Reliability : 3.339
Sequence
CDS
ATGGCAAAAGCACCCGCAGTAGGAATCGATCTGGGTACCACGTACTCTTGCGTTGGTGTCTTCCAGCACGGGAAGGTGGAGATCATCGCCAACGACCAGGGCAACAGGACCACTCCGTCTTATGTTGCGTTCACAGACACCGAGCGTCTCATCGGAGATGCCGCCAAGAACCAGGTGGCGATGAACCCCAACAACACAATATTCGATGCCAAACGTCTCATCGGACGTAAGTTCGAAGATGCTACTGTGCAAGCCGACATGAAGCACTGGCCTTTCGAGGTTGTCAGTGATGGAGGCAAACCTAAGATCAAGGTAGCATATAAGGGTGAAGACAAAACCTTTTTCCCCGAGGAAGTCAGTTCCATGGTGCTTACGAAAATGAAGGAAACTGCCGAAGCTTATCTTGGCAAAACTGTGCAGAATGCAGTTATCACGGTTCCCGCGTACTTCAATGACTCTCAAAGACAAGCCACAAAAGATGCAGGTACCATCTCTGGCTTGAACGTTCTCCGAATCATCAATGAACCGACTGCTGCTGCGATTGCTTACGGTCTTGACAAAAAGGGTACTGGAGAACGAAATGTACTTATCTTTGACCTCGGCGGCGGTACCTTCGACGTGTCCATCCTTACCATCGAGGATGGTATCTTCGAGGTGAAATCCACCGCCGGCGACACCCACTTGGGAGGTGAGGACTTTGACAATCGCATGGTCAACCACTTTGTCCAGGAGTTCAAGAGGAAATACAAAAAGGACCTCGCTACCAACAAGAGAGCTCTTAGGCGTTTGCGTACTGCATGTGAGAGGGCAAAGAGGACCTTGTCATCGTCCACACAAGCGAGCATTGAGATAGATTCTCTCTTTGAGGGTATTGACTTCTACACGTCAATTACTCGTGCTCGCTTCGAGGAGCTGAACGCCGATCTGTTCAGGTCTACCATGGAGCCAGTGGAGAAGTCTCTCCGTGATGCCAAGATGGATAAGGCTCAAATCCACGATATTGTACTGGTGGGTGGCTCCACTCGTATCCCCAAGGTGCAGAAGCTCCTGCAAGATTTCTTTAATGGAAAGGAGCTCAACAAATCTATTAACCCTGACGAGGCCGTAGCTTATGGTGCAGCTGTCCAGGCTGCTATCTTGCACGGTGACAAGTCTGAGGAGGTGCAGGATCTGCTGTTGCTTGATGTAACACCCCTTTCCCTCGGTATTGAGACTGCTGGAGGTGTCATGACCACACTCATCAAGCGTAACACTACCATCCCCACTAAACAGACTCAGACATTCACCACCTACTCTGATAACCAACCCGGAGTACTCATCCAAGTATTTGAGGGTGAGCGTGCTATGACCAAAGATAACAACTTGCTCGGTAAATTCGAGCTGACCGGGATCCCACCGGCGCCGCGTGGCGTGCCTCAAATTGAGGTCACCTTCGACATCGATGCCAACGGTATCCTCAACGTTTCCGCTATCGAGAAGTCCACCAACAAGGAGAACAAGATCACCATTACCAACGACAAAGGTCGTCTCTCCAAGGAAGAGATCGAGCGTATGGTTAATGAGGCAGAGAAGTACAGAAACGAGGATGACAAGCAAAAGGAGACCATCCAGGCCAAGAATGCATTGGAATCTTACTGCTTCAGCATGAAGTCTACCATGGAGGATGAGAAGCTCAAGGAAAAGATCTCTGACTCTGACAAGCAGACCATCCTCGACAAGTGCAACGACACCATCAAGTGGCTGGATTCCAACCAGCTGGCCGACAAGGAGGAGTATGAGCACAAGCAGAAAGAATTGGAAGGCATTTACAATCCGATAATTACGAAGATGTACCAGGGTGCCGGAGGAGTCCCCGGAGGTATGCCGGGCTTCCCGGGCGGAGCACCCGGAGCCGGAGGTGCCGCCCCCGGGGCTGGAGGCGCTGGCCCCACCATCGAGGAGGTCGATTAA
Protein
MAKAPAVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQVAMNPNNTIFDAKRLIGRKFEDATVQADMKHWPFEVVSDGGKPKIKVAYKGEDKTFFPEEVSSMVLTKMKETAEAYLGKTVQNAVITVPAYFNDSQRQATKDAGTISGLNVLRIINEPTAAAIAYGLDKKGTGERNVLIFDLGGGTFDVSILTIEDGIFEVKSTAGDTHLGGEDFDNRMVNHFVQEFKRKYKKDLATNKRALRRLRTACERAKRTLSSSTQASIEIDSLFEGIDFYTSITRARFEELNADLFRSTMEPVEKSLRDAKMDKAQIHDIVLVGGSTRIPKVQKLLQDFFNGKELNKSINPDEAVAYGAAVQAAILHGDKSEEVQDLLLLDVTPLSLGIETAGGVMTTLIKRNTTIPTKQTQTFTTYSDNQPGVLIQVFEGERAMTKDNNLLGKFELTGIPPAPRGVPQIEVTFDIDANGILNVSAIEKSTNKENKITITNDKGRLSKEEIERMVNEAEKYRNEDDKQKETIQAKNALESYCFSMKSTMEDEKLKEKISDSDKQTILDKCNDTIKWLDSNQLADKEEYEHKQKELEGIYNPIITKMYQGAGGVPGGMPGFPGGAPGAGGAAPGAGGAGPTIEEVD
Summary
Similarity
Belongs to the heat shock protein 70 family.
Keywords
ATP-binding
Nucleotide-binding
Nucleus
Stress response
Feature
chain Heat shock 70 kDa protein cognate 4
Uniprot
Q8N0P2
Q9U639
A0A0A7HBV4
K9LT17
A0A0A7HBC8
S5FXH1
+ More
E3VS41 A0A2D1CSD1 T1SC35 F6MFD1 A0A2H1WFA2 Q4ZJ79 E0XN32 R4V6T6 Q5MGD5 A0A343EU24 E2F395 Q0MUU8 A0A2H4LI91 A2TEL6 A2TLM5 A0A2A4IV01 B0FC98 Q0KKB3 C8CCR4 A0A212EUW9 A2TE70 E5LEV4 Q94805 C7SIR9 D5KZX7 J9PWW6 A0A194QIV8 A0A0L7L284 I4DQY2 A0A0N1II90 Q2WG65 A0A023RDH9 K7NAP5 Q3V6C5 A5A3D7 A0A060AEV4 D2KCJ3 A3FKF1 A2TE71 T1YSQ6 A0A075DZG5 T1HJT8 R4G5M1 A0A023F9S2 K9L9P4 E2AKV6 E0V9N7 A0A2R7WZB9 A0A1I9WLD8 A0A069DWP1 N6UDC1 M1KYK4 D5KXW8 U4UVV8 A0A139WAS5 S5FYN8 A0A195DMR5 A0A1W4XB25 A0A109UU25 R4WCP9 A0A0U2PPY2 A0A1Y1ML84 A0A077EU92 A0A1S5VZF6 A0A151IKI9 B3TNP0 A0A026WAP1 A0A060A9H7 A0A0J7KCL5 A9XG01 J7FTR8 F4W666 A0A2A3EG18 V9IEZ2 A0A0M9A1H2 A0A087ZQ27 A0A195FKD0 A0A346THM7 A0A195AWF7 A0A158P1Z0 A0A158P1Z1 K7IQ77 A0A0A9W3D3 A0A0A9VW00 A0A224X8Y3 A0A182FNZ2 A0A182R4T2 A0A0B4KJ04 J9JWP2 U6BQX5 A0A182N5U8 A0A346THN5 A0A1W6EW99 A0A2P0XIF9
E3VS41 A0A2D1CSD1 T1SC35 F6MFD1 A0A2H1WFA2 Q4ZJ79 E0XN32 R4V6T6 Q5MGD5 A0A343EU24 E2F395 Q0MUU8 A0A2H4LI91 A2TEL6 A2TLM5 A0A2A4IV01 B0FC98 Q0KKB3 C8CCR4 A0A212EUW9 A2TE70 E5LEV4 Q94805 C7SIR9 D5KZX7 J9PWW6 A0A194QIV8 A0A0L7L284 I4DQY2 A0A0N1II90 Q2WG65 A0A023RDH9 K7NAP5 Q3V6C5 A5A3D7 A0A060AEV4 D2KCJ3 A3FKF1 A2TE71 T1YSQ6 A0A075DZG5 T1HJT8 R4G5M1 A0A023F9S2 K9L9P4 E2AKV6 E0V9N7 A0A2R7WZB9 A0A1I9WLD8 A0A069DWP1 N6UDC1 M1KYK4 D5KXW8 U4UVV8 A0A139WAS5 S5FYN8 A0A195DMR5 A0A1W4XB25 A0A109UU25 R4WCP9 A0A0U2PPY2 A0A1Y1ML84 A0A077EU92 A0A1S5VZF6 A0A151IKI9 B3TNP0 A0A026WAP1 A0A060A9H7 A0A0J7KCL5 A9XG01 J7FTR8 F4W666 A0A2A3EG18 V9IEZ2 A0A0M9A1H2 A0A087ZQ27 A0A195FKD0 A0A346THM7 A0A195AWF7 A0A158P1Z0 A0A158P1Z1 K7IQ77 A0A0A9W3D3 A0A0A9VW00 A0A224X8Y3 A0A182FNZ2 A0A182R4T2 A0A0B4KJ04 J9JWP2 U6BQX5 A0A182N5U8 A0A346THN5 A0A1W6EW99 A0A2P0XIF9
Pubmed
EMBL
BABH01011152
HQ694956
AB084922
ADW41775.1
BAB92074.1
AF194819
+ More
KJ639908 AIZ00748.1 JQ340172 AFO70210.1 KJ639909 AIZ00749.1 KC844151 KF731994 AGQ50302.1 AHF81958.1 HQ223341 ADO32621.1 KX977589 ATN45248.1 JX680673 JX680674 AGR84220.1 AGR84221.1 JF809817 AEG78288.1 ODYU01008280 SOQ51731.1 DQ004584 AAY26452.2 GU433378 ADM66138.1 KC787696 AGM34286.1 AY829851 AAV91465.1 MF067297 ASM61895.1 HM046611 ADK55518.1 DQ845103 DQ845104 ABH09733.1 KX845567 ATB54998.1 EF194276 ABM90551.1 EF213550 ABN09627.1 NWSH01007300 PCG63012.1 EU306518 ABY55233.1 AB251896 BAF03556.1 GQ389712 ACV32641.1 AGBW02012297 OWR45261.1 EF190212 ABM90802.1 HQ434763 JF708083 JF708084 ADR00357.2 AEO19922.1 AEO19923.1 U23504 AAB06239.1 FJ432703 ACL31668.1 GU726137 RSAL01000148 ADE05296.1 RVE45838.1 JN676213 AFC38439.1 KQ458714 KPJ05447.1 JTDY01003535 KOB69384.1 AK404837 BAM20322.1 KQ459934 KPJ19257.1 AB214973 BAE48743.1 KJ437496 AHX58694.1 HQ698836 ADZ15147.1 AB206478 BAE44308.1 EF523379 ABP93403.1 KJ573767 AIA61347.1 GU205816 ADA61012.1 EF392678 ABN50911.1 EF190213 ABM90803.1 KF052087 AGU67952.1 KF311064 AHY95944.1 ACPB03018406 GAHY01000410 JAA77100.1 GBBI01000572 GEMB01001689 JAC18140.1 JAS01474.1 JN811146 AFG29035.1 GL440413 EFN65945.1 DS234995 EEB10106.1 KK856104 PTY24888.1 KU932322 APA33958.1 GBGD01000767 JAC88122.1 APGK01039372 APGK01039373 KB740970 ENN76652.1 KC184398 AGF33487.1 GU723301 ADE34170.1 KB632388 ERL94381.1 KQ971380 KYB25034.1 KC844150 AGQ50301.1 KQ980724 KYN14122.1 KP994340 QKKF02017343 AMD09926.1 RZF40992.1 AK416914 BAN20129.1 KR181952 ALJ76676.1 GEZM01028132 JAV86444.1 KJ909506 AIL52740.1 KX976471 AQP31361.1 KQ977199 KYN04993.1 EU340838 ACA53150.1 KK107293 QOIP01000005 EZA53152.1 RLU22879.1 KJ573768 AIA61348.1 LBMM01009632 KMQ87979.1 EF208962 ABP97091.1 JX112895 AFP54307.1 GL887707 EGI70201.1 KZ288254 PBC30723.1 JR042487 AEY59625.1 KQ435763 KOX75410.1 KQ981523 KYN40454.1 MG265904 AXU24960.1 KQ976731 KYM76299.1 ADTU01006918 GBHO01044229 JAF99374.1 GBHO01044228 GBRD01005543 JAF99375.1 JAG60278.1 GFTR01007630 JAW08796.1 KC119044 AGM20720.1 ABLF02026083 KC544268 AHA36968.1 MG265912 AXU24968.1 KY563591 ARK20000.1 KY661334 AVA17421.1
KJ639908 AIZ00748.1 JQ340172 AFO70210.1 KJ639909 AIZ00749.1 KC844151 KF731994 AGQ50302.1 AHF81958.1 HQ223341 ADO32621.1 KX977589 ATN45248.1 JX680673 JX680674 AGR84220.1 AGR84221.1 JF809817 AEG78288.1 ODYU01008280 SOQ51731.1 DQ004584 AAY26452.2 GU433378 ADM66138.1 KC787696 AGM34286.1 AY829851 AAV91465.1 MF067297 ASM61895.1 HM046611 ADK55518.1 DQ845103 DQ845104 ABH09733.1 KX845567 ATB54998.1 EF194276 ABM90551.1 EF213550 ABN09627.1 NWSH01007300 PCG63012.1 EU306518 ABY55233.1 AB251896 BAF03556.1 GQ389712 ACV32641.1 AGBW02012297 OWR45261.1 EF190212 ABM90802.1 HQ434763 JF708083 JF708084 ADR00357.2 AEO19922.1 AEO19923.1 U23504 AAB06239.1 FJ432703 ACL31668.1 GU726137 RSAL01000148 ADE05296.1 RVE45838.1 JN676213 AFC38439.1 KQ458714 KPJ05447.1 JTDY01003535 KOB69384.1 AK404837 BAM20322.1 KQ459934 KPJ19257.1 AB214973 BAE48743.1 KJ437496 AHX58694.1 HQ698836 ADZ15147.1 AB206478 BAE44308.1 EF523379 ABP93403.1 KJ573767 AIA61347.1 GU205816 ADA61012.1 EF392678 ABN50911.1 EF190213 ABM90803.1 KF052087 AGU67952.1 KF311064 AHY95944.1 ACPB03018406 GAHY01000410 JAA77100.1 GBBI01000572 GEMB01001689 JAC18140.1 JAS01474.1 JN811146 AFG29035.1 GL440413 EFN65945.1 DS234995 EEB10106.1 KK856104 PTY24888.1 KU932322 APA33958.1 GBGD01000767 JAC88122.1 APGK01039372 APGK01039373 KB740970 ENN76652.1 KC184398 AGF33487.1 GU723301 ADE34170.1 KB632388 ERL94381.1 KQ971380 KYB25034.1 KC844150 AGQ50301.1 KQ980724 KYN14122.1 KP994340 QKKF02017343 AMD09926.1 RZF40992.1 AK416914 BAN20129.1 KR181952 ALJ76676.1 GEZM01028132 JAV86444.1 KJ909506 AIL52740.1 KX976471 AQP31361.1 KQ977199 KYN04993.1 EU340838 ACA53150.1 KK107293 QOIP01000005 EZA53152.1 RLU22879.1 KJ573768 AIA61348.1 LBMM01009632 KMQ87979.1 EF208962 ABP97091.1 JX112895 AFP54307.1 GL887707 EGI70201.1 KZ288254 PBC30723.1 JR042487 AEY59625.1 KQ435763 KOX75410.1 KQ981523 KYN40454.1 MG265904 AXU24960.1 KQ976731 KYM76299.1 ADTU01006918 GBHO01044229 JAF99374.1 GBHO01044228 GBRD01005543 JAF99375.1 JAG60278.1 GFTR01007630 JAW08796.1 KC119044 AGM20720.1 ABLF02026083 KC544268 AHA36968.1 MG265912 AXU24968.1 KY563591 ARK20000.1 KY661334 AVA17421.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000037510
+ More
UP000053240 UP000015103 UP000000311 UP000009046 UP000019118 UP000030742 UP000007266 UP000078492 UP000192223 UP000291343 UP000078542 UP000053097 UP000279307 UP000036403 UP000007755 UP000242457 UP000053105 UP000005203 UP000078541 UP000078540 UP000005205 UP000002358 UP000069272 UP000075900 UP000007819 UP000075884
UP000053240 UP000015103 UP000000311 UP000009046 UP000019118 UP000030742 UP000007266 UP000078492 UP000192223 UP000291343 UP000078542 UP000053097 UP000279307 UP000036403 UP000007755 UP000242457 UP000053105 UP000005203 UP000078541 UP000078540 UP000005205 UP000002358 UP000069272 UP000075900 UP000007819 UP000075884
Interpro
IPR029047
HSP70_peptide-bd_sf
+ More
IPR013126 Hsp_70_fam
IPR018181 Heat_shock_70_CS
IPR029048 HSP70_C_sf
IPR019395 Transmembrane_161A/B
IPR014753 Arrestin_N
IPR011021 Arrestin-like_N
IPR014752 Arrestin_C
IPR014756 Ig_E-set
IPR011022 Arrestin_C-like
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR013126 Hsp_70_fam
IPR018181 Heat_shock_70_CS
IPR029048 HSP70_C_sf
IPR019395 Transmembrane_161A/B
IPR014753 Arrestin_N
IPR011021 Arrestin-like_N
IPR014752 Arrestin_C
IPR014756 Ig_E-set
IPR011022 Arrestin_C-like
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
Gene 3D
ProteinModelPortal
Q8N0P2
Q9U639
A0A0A7HBV4
K9LT17
A0A0A7HBC8
S5FXH1
+ More
E3VS41 A0A2D1CSD1 T1SC35 F6MFD1 A0A2H1WFA2 Q4ZJ79 E0XN32 R4V6T6 Q5MGD5 A0A343EU24 E2F395 Q0MUU8 A0A2H4LI91 A2TEL6 A2TLM5 A0A2A4IV01 B0FC98 Q0KKB3 C8CCR4 A0A212EUW9 A2TE70 E5LEV4 Q94805 C7SIR9 D5KZX7 J9PWW6 A0A194QIV8 A0A0L7L284 I4DQY2 A0A0N1II90 Q2WG65 A0A023RDH9 K7NAP5 Q3V6C5 A5A3D7 A0A060AEV4 D2KCJ3 A3FKF1 A2TE71 T1YSQ6 A0A075DZG5 T1HJT8 R4G5M1 A0A023F9S2 K9L9P4 E2AKV6 E0V9N7 A0A2R7WZB9 A0A1I9WLD8 A0A069DWP1 N6UDC1 M1KYK4 D5KXW8 U4UVV8 A0A139WAS5 S5FYN8 A0A195DMR5 A0A1W4XB25 A0A109UU25 R4WCP9 A0A0U2PPY2 A0A1Y1ML84 A0A077EU92 A0A1S5VZF6 A0A151IKI9 B3TNP0 A0A026WAP1 A0A060A9H7 A0A0J7KCL5 A9XG01 J7FTR8 F4W666 A0A2A3EG18 V9IEZ2 A0A0M9A1H2 A0A087ZQ27 A0A195FKD0 A0A346THM7 A0A195AWF7 A0A158P1Z0 A0A158P1Z1 K7IQ77 A0A0A9W3D3 A0A0A9VW00 A0A224X8Y3 A0A182FNZ2 A0A182R4T2 A0A0B4KJ04 J9JWP2 U6BQX5 A0A182N5U8 A0A346THN5 A0A1W6EW99 A0A2P0XIF9
E3VS41 A0A2D1CSD1 T1SC35 F6MFD1 A0A2H1WFA2 Q4ZJ79 E0XN32 R4V6T6 Q5MGD5 A0A343EU24 E2F395 Q0MUU8 A0A2H4LI91 A2TEL6 A2TLM5 A0A2A4IV01 B0FC98 Q0KKB3 C8CCR4 A0A212EUW9 A2TE70 E5LEV4 Q94805 C7SIR9 D5KZX7 J9PWW6 A0A194QIV8 A0A0L7L284 I4DQY2 A0A0N1II90 Q2WG65 A0A023RDH9 K7NAP5 Q3V6C5 A5A3D7 A0A060AEV4 D2KCJ3 A3FKF1 A2TE71 T1YSQ6 A0A075DZG5 T1HJT8 R4G5M1 A0A023F9S2 K9L9P4 E2AKV6 E0V9N7 A0A2R7WZB9 A0A1I9WLD8 A0A069DWP1 N6UDC1 M1KYK4 D5KXW8 U4UVV8 A0A139WAS5 S5FYN8 A0A195DMR5 A0A1W4XB25 A0A109UU25 R4WCP9 A0A0U2PPY2 A0A1Y1ML84 A0A077EU92 A0A1S5VZF6 A0A151IKI9 B3TNP0 A0A026WAP1 A0A060A9H7 A0A0J7KCL5 A9XG01 J7FTR8 F4W666 A0A2A3EG18 V9IEZ2 A0A0M9A1H2 A0A087ZQ27 A0A195FKD0 A0A346THM7 A0A195AWF7 A0A158P1Z0 A0A158P1Z1 K7IQ77 A0A0A9W3D3 A0A0A9VW00 A0A224X8Y3 A0A182FNZ2 A0A182R4T2 A0A0B4KJ04 J9JWP2 U6BQX5 A0A182N5U8 A0A346THN5 A0A1W6EW99 A0A2P0XIF9
PDB
5FPN
E-value=0,
Score=2413
Ontologies
PATHWAY
GO
PANTHER
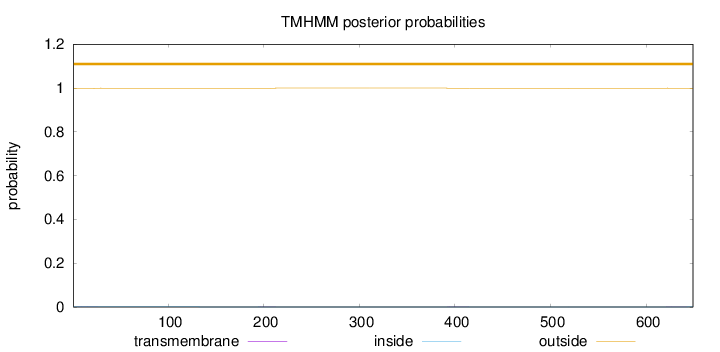
Topology
Subcellular location
Nucleus
Length:
649
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02999
Exp number, first 60 AAs:
0.01495
Total prob of N-in:
0.00137
outside
1 - 649
Population Genetic Test Statistics
Pi
175.266163
Theta
173.958734
Tajima's D
-0.719767
CLR
0.163738
CSRT
0.1978401079946
Interpretation
Uncertain
