Gene
KWMTBOMO05348 Validated by peptides from experiments
Annotation
nimrod_B_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.612
Sequence
CDS
ATGTTGATTATAATATATGTGCTCGGGCTCTTGCTGGTATCCACCGGCGGTCAGTACGCGAAGTCGGGGGTCAAAGGTGGCTACGGAGGTCGCGGCTTCCAAGATAACACCAATGAGAACATCGCCAAATTTTACGGGAACAACACCAGCCATGGTTTTCTATCGCAAAACCATACGGTTAATACCATGAGCGATATCGGAGTATGTTATATCGAAGTACCTACAGCTCAACTTGTCGAGGATCAGAAGCAGATACCTAAAGGAAACGGATCCCGGCCCGATCTCAGCGTCATTCGTTCTTGCTGCAATGGCTACGTGAGGAACATCTACAACCCTAACATTTGCGAGCCGCACTGCAGCAAAGGATGCGTCAACGCATTGTGCACAGCGCCTGAAATCTGCACGTGCTTCCCTGACCACGTCAAGAACGCCGGCGGCTTCTGCATCGCTACATGCCCGATTGGATGTCAAAATGGCCACTGCTCCGGTCGAGAGTGCGTCTGCAGGGATGGCTTTAAACTAGACTACGGACGTAAATACTGTGTGCCAGCTTGCAGTAACAACTGCGCCGGTGTCGGTAATTGCACTTCACCAAACCGATGCGACTGCGCACCTGGTTATCAGGCAACACACGATGGATCGTGCAGTCCACAATGTCGCGATTGCGCGCCCGATGCTTGTGTTGCTCCAAATGTGTGCTCAGGCCCTACCAGAATTCCGTTGCCGGGACAAAACACCCAAATCCCTACATCTAATTATCCAGGATATCATAGTTCCACTTTTTACAATCGTCCAGGGATCGCACAAGGTCCTCTTCCCACCCAACAGCCGAATTATGTTGTCGGTCCCTCATACGGAAGCAGCCAACCAGTTTCTGATAATCAACTAAATGGTCCTTTACACAACACTAGTTTATATCCTTCTTATTCGTTGCAACAATTGAACAATAGCTATCCAGGGCAACAGCCTCAGTATCCTCTACCCAACCAGCCTCAATATCCTTGGCAGCAAAACCCGCATTCCAATCAGCCAAACTACACCTTATTGAATAGCACTCTACACCCACATTGGAAAAATAATTACTTTGATTCCAATCCATCACCATATCAACCACCATATCCAGGACCATTACCGAGCCCGGGCCAACAGCCCATTTATCCAACACAACAGTTTAATCAAACTCAAATGCCACCTCACTATCCTAATCAAAACGTTTATCCTTATCCTTCTCCACTTCAGCCTTGGTTTCCTGGACAAATACCCGCAAACGGTAACCACACGCAAAACGGCTTCCTGGACTTATCCAATCGCACTGTACCTGGTCAAGGTGGCTTCCCTCCCCATCAAGGCTACTATCCGTATCCGCTTTACCAACCGTCGTATCCTCAACCAGGATATTTTCAGCCCAATCAATCCAACTCCGGATACTATGGGCAGCAACCAACACCAAGCACAGATTCCTTACATGCGGGCTATTATAACAATTATGACGGGTCAAATCCTCAGGCGCCTGTATGCTCTCAGCCTTGTCTGAACGGAGTCTGTGTCGAAGGCAACAGATGCTCCTGCAACACTGGATATGTCACTGATAGTATGGACCCCAGTGGATTTAGGTGCATACCTCACTGTGCCGGAGGGTGTCCTAATGGTGTCTGCTCGGCACCAAACCTGTGCATCTGTAACATGGGCTATAAAAAGGACACTAGCGTCAAGGGCAGATCTGTATGCGTCAAGAAAATCAGATGA
Protein
MLIIIYVLGLLLVSTGGQYAKSGVKGGYGGRGFQDNTNENIAKFYGNNTSHGFLSQNHTVNTMSDIGVCYIEVPTAQLVEDQKQIPKGNGSRPDLSVIRSCCNGYVRNIYNPNICEPHCSKGCVNALCTAPEICTCFPDHVKNAGGFCIATCPIGCQNGHCSGRECVCRDGFKLDYGRKYCVPACSNNCAGVGNCTSPNRCDCAPGYQATHDGSCSPQCRDCAPDACVAPNVCSGPTRIPLPGQNTQIPTSNYPGYHSSTFYNRPGIAQGPLPTQQPNYVVGPSYGSSQPVSDNQLNGPLHNTSLYPSYSLQQLNNSYPGQQPQYPLPNQPQYPWQQNPHSNQPNYTLLNSTLHPHWKNNYFDSNPSPYQPPYPGPLPSPGQQPIYPTQQFNQTQMPPHYPNQNVYPYPSPLQPWFPGQIPANGNHTQNGFLDLSNRTVPGQGGFPPHQGYYPYPLYQPSYPQPGYFQPNQSNSGYYGQQPTPSTDSLHAGYYNNYDGSNPQAPVCSQPCLNGVCVEGNRCSCNTGYVTDSMDPSGFRCIPHCAGGCPNGVCSAPNLCICNMGYKKDTSVKGRSVCVKKIR
Summary
Uniprot
Pubmed
Proteomes
PRIDE
Pfam
PF02363 C_tripleX
ProteinModelPortal
PDB
2YGQ
E-value=3.85694e-05,
Score=114
Ontologies
GO
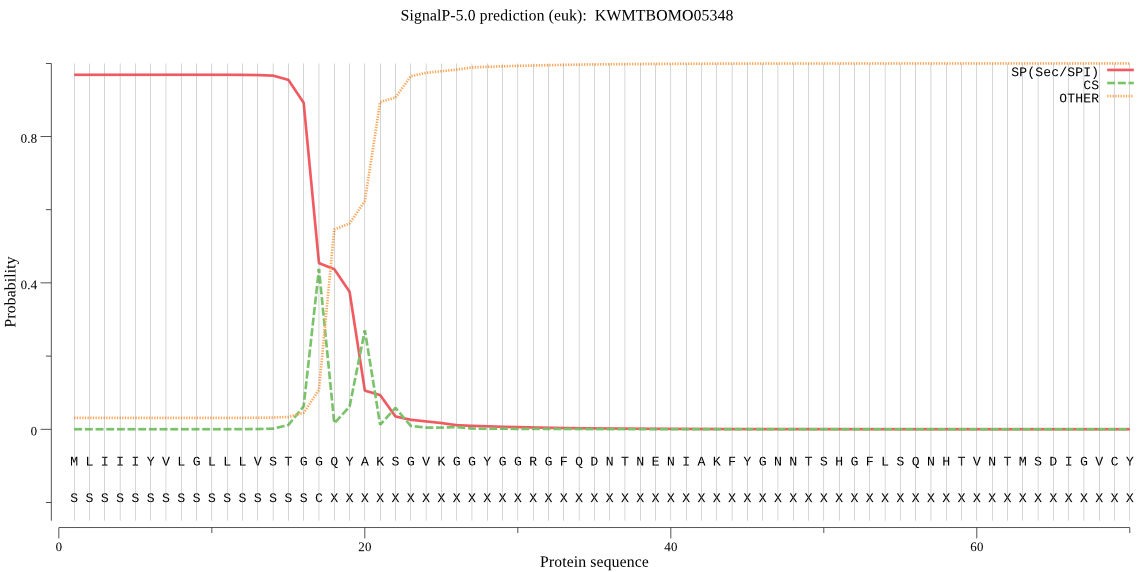
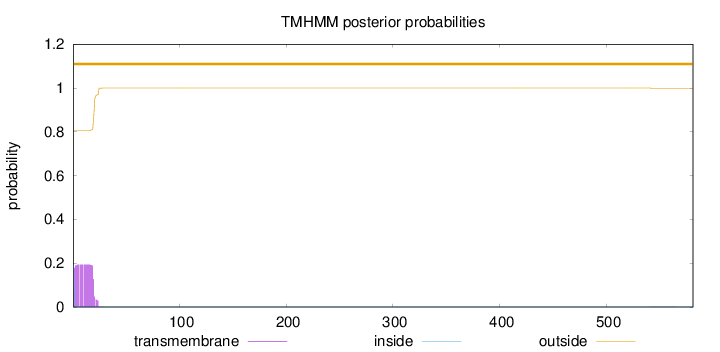
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.968749
Length:
581
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.73423
Exp number, first 60 AAs:
3.72694
Total prob of N-in:
0.19375
outside
1 - 581
Population Genetic Test Statistics
Pi
165.306462
Theta
190.087112
Tajima's D
-1.034182
CLR
1.128942
CSRT
0.130893455327234
Interpretation
Uncertain
