Gene
KWMTBOMO05347
Annotation
hypothetical_protein_[Mycobacterium_yongonense]
Location in the cell
Extracellular Reliability : 1.888 Nuclear Reliability : 1.528
Sequence
CDS
ATGAAGATTTGTGAAATAAAGAAGCGGTGTGGTGGTACCTACCTGTCTGTTTCTACAAAACTACCTGGGCTTGTTTTTAGATTTCTGGTCTTTGATCCCGCTGCTCCTGTCCTGCCTGTAGGTCCTGTCTGTGGTCTAGTAGGTTCTGGCTGTGCTCCAGTTGATTCTGGGTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGTAAATCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGGTCTGGCTGTGGTCTAGTAAGTTCTGGCTGTGCTCCAGTAAGTCTTGTCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGAGTTCTGGCTGTTGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTAGGTTCTGGCTGTTGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTATTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTAATTCTGGTTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTATTAAGTTCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCTAGTAAGTCCTGGGTGTGCTCCAGTTGATTCTGGCTGTGGTCCGGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGTTGTGCTCCAATTGATTCTGGCTTAGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGTTGTGCTCCAGTTGATTCTGATTGTGCTGTAGAGGGTTCTGGCTGTGCTGCAGTGGGTTCTGGCTGTGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTATCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAGTCCTGACTGTGCTCTAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTGCTCCAGTAAGTCCTGGCTGTGCCCCAGTTGATTCTGGCTGTGGTCTAGTAAGTCTTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAGTCCTGAGTGTGCTCCAGTTGATTTTGGCTGTGTTGCAGTGGGTTCAGGCTGTGCTCCAGTAAGTCCTGGCTGTGCCCTAGTTGATTCTGGCTGTGATCTAGAAAGTCCTGGCTGTGCTCTAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTTTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTATGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGTTGTGGTCCAGTAGATTCTGGCTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTTTGGCTGTGTTGCAGTGGGTTCTGATTGTTGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTTTGGCTGTGTTGCAGTGGGTTCTGATTGTTGTCTAGTAAGCCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGTTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTATGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGTTGTGCACCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAATTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGGTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCATTGGTATCTGGCTATGGTCTAGTAAGTCCTGGTTGTGCACCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGGTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGATTGTTGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGTTGTGCACCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGGTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGATTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCACCAGTTGATTCTGGCTGTGTCGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGATTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTTTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGTTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTATGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCTGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGTTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAATCCTGACTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGTTGTTGTCTAGAAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGGTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGCTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGAGTTCTGGCTGTTGTCTAGTAAGTCCTGACTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGTATCTGGCTATGGTCTAGTAAGTCCTGGTCTTGCACCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCTGGCTGTTGTCTAGGAAGTCCTGGCTGTGCTCCAGTAGATTCTGGCTGTGGTCTAGTAAGTCCTGGTTGTGCTCCAGTTGATTCTGGCTGTGTTGCAGTGGGTTCCGGCTGTGGTCTAAAAGTCCTGTCTGTGGTGCAGTAG
Protein
MKICEIKKRCGGTYLSVSTKLPGLVFRFLVFDPAAPVLPVGPVCGLVGSGCAPVDSGCVAVGSGCCLVNPGCAPVDSGCVAVGSGCGLVSSGCAPVSLVCAPVDSGCVAVSSGCCLVSPGCAPVDSGCVAVGSGCCLVSPGCAPVDSGCVAVGSGCCLLSPGCAPVDSGCGLVSPGCAPVNSGCGLVSPGCAPVDSGCVAVGSGCCLLSSGCAPVDSGCGLVSPGCAPVDSGCGPVDSGCGLVSPGCAPIDSGLGPVDSGCGLVSPGCAPVDSDCAVEGSGCAAVGSGCVDSGCVAVGSGCYLESPGCAPVDSGCVAVVSGYGLGSPDCAPVDSGCVAVGSGCCLGSPDCALVDSGCVAVGSGCAPVSPGCAPVDSGCGLVSLGCAPVDSGCVAVGSGCCLGSPECAPVDFGCVAVGSGCAPVSPGCALVDSGCDLESPGCALVDSGCVAVGSGCFLESPGCAPVDSGYVAVGSGCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCGPVDSGCGPVDSGCGLVSPGCAPVDFGCVAVGSDCCLVSPGCAPVDFGCVAVGSDCCLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDYGCVAVVSGYGLVSPGCAPVDSGCVAVGSGCCLEIPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCVAVGSGCGLVSPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCVAVVSGYGLGSPDCAPVDSGCVAVGSGCCLGSPDCAPVDSGCVALVSGYGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCVAVGSGCGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVGSDCCLGSPDCAPVDSGCVAVVSGYGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCVAVGSGCGLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVGSDCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVGSDCCLESPGCAPVDSGCVAVVSGYGLGSPDCAPVDSGCVAVGSGCCLGSPDCAPVDSGCVAVVSGCGLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVGSGCFLESPGCAPVDSGCGPVDSGCGLVSPGCAPVDYGCVAVGSGCCLGSPDCAPVDSGCGLVSPGCAPVDSGCVAVGSGCCLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDSGCVAVGSGCCLGNPDCGPVDSGCGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDSGCVAVGSGCCLESPGCAPVDSGCVAVVSGYGLVSPGCAPVDSGCGPVDSGCGLVSPGCAPVDSGCVAVSSGCCLVSPDCAPVDSGCVAVVSGYGLVSPGLAPVDSGCVAVGSGCCLGSPGCAPVDSGCGLVSPGCAPVDSGCVAVGSGCGLKVLSVVQ
Summary
Similarity
Belongs to the CRISP family.
Uniprot
EMBL
CP003491
AFJ36544.1
CP003347
AGP65182.1
MVIJ01000021
ORB73291.1
+ More
FUFA01000005 SPM37153.1 NCXM01000068 OSC19304.1 LZSC01000063 OBA58946.1 NEDP02001212 OWF53796.1 FNKX01000002 SDR55567.1 FXEG02000002 SOX53164.1 NBSQ01000002 PNE76469.1 LZIZ01000077 OBG66985.1 LZSC01000081 OBA57650.1 JARK01001352 EYC23112.1 KN737008 KIH55634.1 PNEO01000009 PLZ02360.1 PNEO01000028 PLZ00274.1 KI657551 ETN86459.1
FUFA01000005 SPM37153.1 NCXM01000068 OSC19304.1 LZSC01000063 OBA58946.1 NEDP02001212 OWF53796.1 FNKX01000002 SDR55567.1 FXEG02000002 SOX53164.1 NBSQ01000002 PNE76469.1 LZIZ01000077 OBG66985.1 LZSC01000081 OBA57650.1 JARK01001352 EYC23112.1 KN737008 KIH55634.1 PNEO01000009 PLZ02360.1 PNEO01000028 PLZ00274.1 KI657551 ETN86459.1
Proteomes
Pfam
Interpro
IPR000030
PPE_family
+ More
IPR038332 PPE_sf
IPR011050 Pectin_lyase_fold/virulence
IPR012332 P22_tailspike-like_C_sf
IPR008619 Filamentous_hemagglutn_rpt
IPR030930 AIDA
IPR003992 Pertactin
IPR005594 YadA_C
IPR024973 ESPR
IPR026737 GOLGA6L
IPR008640 Adhesin_Head_dom
IPR011049 Serralysin-like_metalloprot_C
IPR008635 Coiled_stalk_dom
IPR001283 CRISP-related
IPR014044 CAP_domain
IPR018244 Allrgn_V5/Tpx1_CS
IPR035940 CAP_sf
IPR036709 Autotransporte_beta_dom_sf
IPR005546 Autotransporte_beta
IPR038332 PPE_sf
IPR011050 Pectin_lyase_fold/virulence
IPR012332 P22_tailspike-like_C_sf
IPR008619 Filamentous_hemagglutn_rpt
IPR030930 AIDA
IPR003992 Pertactin
IPR005594 YadA_C
IPR024973 ESPR
IPR026737 GOLGA6L
IPR008640 Adhesin_Head_dom
IPR011049 Serralysin-like_metalloprot_C
IPR008635 Coiled_stalk_dom
IPR001283 CRISP-related
IPR014044 CAP_domain
IPR018244 Allrgn_V5/Tpx1_CS
IPR035940 CAP_sf
IPR036709 Autotransporte_beta_dom_sf
IPR005546 Autotransporte_beta
Gene 3D
ProteinModelPortal
Ontologies
PANTHER
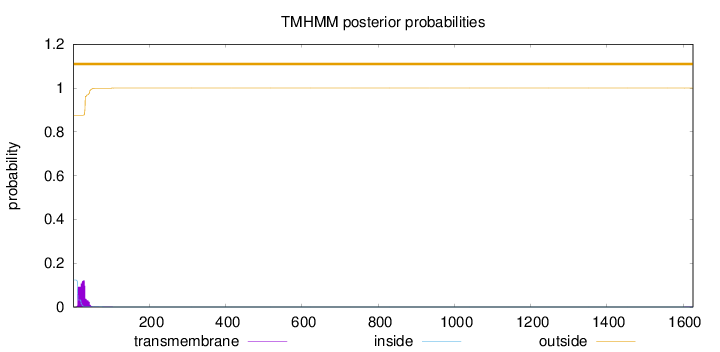
Topology
Length:
1626
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.51502000000001
Exp number, first 60 AAs:
2.48431
Total prob of N-in:
0.12378
outside
1 - 1626
Population Genetic Test Statistics
Pi
179.685664
Theta
169.163482
Tajima's D
-2.13632
CLR
227.468796
CSRT
0.00859957002149892
Interpretation
Uncertain
