Gene
KWMTBOMO05340
Pre Gene Modal
BGIBMGA002528
Annotation
PREDICTED:_angiotensin-converting_enzyme_[Bombyx_mori]
Full name
Angiotensin-converting enzyme
Location in the cell
Nuclear Reliability : 1.931
Sequence
CDS
ATGGCCTGTCTCACTAGGAAGTTTTCCCTATCATGGGTGCAGCTTGGCCTGGTTTTGTGCTTTATCTATTGCGCTATCGGCGTTCCGCAGTTAGACCTACCGCCGATTCCCAGTACTTCTCAAAGCGGCTATGGCCTATCTAGTACTCCATCGTCTCTCAATTCTCCCGGGAATCAGTTTGACAGGAATTATCAGTTCAGCACTGCGAGACCGAGTCCTACCAGCACGCCTGGAGTTTACCAAGTTTCCTCCACCCCGTTCAACGGAATAAGTCCGGATATAAGCAACAATGGGTATCCGAATGTTGACTATAACAGCAGAAACCCTAGTTCTACGCTCCGACCTTATGACGAAAGTAACGGAGACGACAGAATTGACATTAACAGTGATCCGAACTTCAGAAGAAACGACCCCAACTTTGTCAGGAACGACCCGAACTACGTAGGAACTAATCCGTATGATTTCAATCGGGGCGGGGATGTAAATAGTATTGATGATAGATACCGCCAAGTGAACTTGCAGCAGGTCAGAGATTTTCTGGCACAGGCTGACGATCAGGCTTCGAAGGAATGCACCAATAACGTCGCAGCGCAGTGGAACTTCGAAACAAACGTCAATGATGAAACACAGCATGCAGCGCTGGACGCCCAACAGCGCTACACATTGTTCCAGCGAGGACTATGGGAGGCCGCACAGCAAATACCTCGTGGAGCTATCAGGGACTTTCCGACCTTCAGACAACTCCGGCTTCTGTCTATTATAGGACCTGCAGCTCTACCCCCTGATCAACTGGATAGGTACAACCGCATAATAAACGACATGCTGGCTGTTTACAATACTGCCGAGGTCTGCGCCTACAACGAGCCATTCAAGTGTGGTCTCCATCTCCAACCGGAGCTGCAGTACATCATGTCCCACTCCAGAGACTGGGACGAACTTCAGCACATCTGGACCGAGTGGAGGAGGAATACTGGTAGAAGAGTACGGGATTTATACGAGCAGTTGGTTGATTTGACCAATCAGGCAGCCAGATTGAATAATTTTACTGACGCATCCGCGTACTGGATGTTTCCTTACGAGACCTCAAACATGCGGCAGGAGGTCGATGACGTCTGGCAGCAGATCAAGCCCCTTTATGACCAGCTTCACGCCTACGTTCGTCGTCGTCTGCGAGAAGCCTACGGTCCTGAGAGAATCTCCAGATCGGCTCCGTTACCAGCCCACATCCTGGGAGACATGTGGGGACAGAGCTGGTCGGGTATCGTGCCTTTTACCCTTCCTTACCCTGGGAAAAACTTGCTAGATGTCTCTCAGGAAATGGCGAAGCAGGGTTTCACTCCACTCACAATGTTTCAATTAGCTGAAGAGTACTTCGTGTCCATGAATATGTCTGCCATGCCTCCTGACTTTTGGGCCCTAAGCGTGTTGGACCAACCTCCGGACAGACACATCCACTGCCAGCCTTCTGCTTGGGACTTTTGCAATAAGCACGACTATCGCATCAAAATGTGCACACACGTGGACATGAAGGATCTGGTGACGGCGCATCATGAGATGGCGCACATCGAATATTTCCTCGCCTACAGGAATCAGCCGAAAGTCTTCCGTGATGGTGCTAATCCAGGTTTCCACGAAGCTATTGGTGAGACAATAGCACTGTCAGTTGGATCCCCTCGTCACCTGCAAACCCTAGGCCTGGTTCAACGTTCGGTAGACGACACAGCCCACAACACGAACTTCCTGTTCACCCAAGCTATGGACAAGTTAGCGTTTCTTCCGTTCGCGCTGGTGATGGACAGGTGGCGTTGGGACGTGTTCACAGGAGACGTCCGCAAGGAGCAGTACAATTGCCATTGGTGGAGATTAAGGGAGCAGTACCAAGGTGTGAAGCCTCCCGTGCTGCGTTCCGAGCTGGACTTCGACCCTGGATCCAAGTACCACATACCGGCCAACATACCCTACATAAGGTCAGTTTAG
Protein
MACLTRKFSLSWVQLGLVLCFIYCAIGVPQLDLPPIPSTSQSGYGLSSTPSSLNSPGNQFDRNYQFSTARPSPTSTPGVYQVSSTPFNGISPDISNNGYPNVDYNSRNPSSTLRPYDESNGDDRIDINSDPNFRRNDPNFVRNDPNYVGTNPYDFNRGGDVNSIDDRYRQVNLQQVRDFLAQADDQASKECTNNVAAQWNFETNVNDETQHAALDAQQRYTLFQRGLWEAAQQIPRGAIRDFPTFRQLRLLSIIGPAALPPDQLDRYNRIINDMLAVYNTAEVCAYNEPFKCGLHLQPELQYIMSHSRDWDELQHIWTEWRRNTGRRVRDLYEQLVDLTNQAARLNNFTDASAYWMFPYETSNMRQEVDDVWQQIKPLYDQLHAYVRRRLREAYGPERISRSAPLPAHILGDMWGQSWSGIVPFTLPYPGKNLLDVSQEMAKQGFTPLTMFQLAEEYFVSMNMSAMPPDFWALSVLDQPPDRHIHCQPSAWDFCNKHDYRIKMCTHVDMKDLVTAHHEMAHIEYFLAYRNQPKVFRDGANPGFHEAIGETIALSVGSPRHLQTLGLVQRSVDDTAHNTNFLFTQAMDKLAFLPFALVMDRWRWDVFTGDVRKEQYNCHWWRLREQYQGVKPPVLRSELDFDPGSKYHIPANIPYIRSV
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M2 family.
Feature
chain Angiotensin-converting enzyme
Uniprot
A0A212F3F9
A0A3S2TLH5
A0A2A4IVE5
A0A0N1IC85
A0A194QJ89
A0A182NBR7
+ More
A0A2M4CGD1 W5JF67 A0A1Y9G9G1 A0A2M4BCK2 A0A2M4BCM7 A0A2M4BDM0 A0A182R3U4 A0A182Y0N9 A0A2M3Z1D4 A0A2M3ZE07 A0A182WGJ6 A0A182QWK5 A0A182K2I1 A0A182XN91 A0A182LZL0 A0A182JJ88 A0A182U8D7 A0A182UXG9 A0A240PL25 A0A182HRX0 Q7Q1J9 A0A182GI45 A0A1B6DUB7 A0A182GQM7 Q17HJ9 A0A182KU23 A0A1B6KA09 A0A1B6GNU4 A0A2J7QU96 A0A139WAY6 B0WR91 A0A1Y1M039 U4TXW0 A0A1J1IC50 A0A1B0GNY6 A0A084WU13 A0A2K9Y420 A0A2H8TCU4 A0A2S2PGF5 J9K345 A0A2S2PBL3 A0A2S2PXK3 A0A088AG98 A0A3Q0IXC2 A0A026VXF5 A0A3L8DDW7 E2CAC6 E0VYZ7 A0A158P0W6 A0A154PMJ3 A0A336N2A5 A0A336MZK1 A0A0C9RBF0 A0A0K8T5T2 A0A195CXY5 A0A1B0CG74 A0A0L7QVQ3 A0A310SFD5 A0A182SJM2 A0A023F3X6 K7IXA0 A0A151WW54 A0A0L0BM21 A0A1A9WU17 A0A1I8QEP7 Q8SXX2 B3MN04 B3NAA9 B4JCZ2 A0A3B0JNP4 B5DJZ6 B4HXC1 A0A0J9R277 A0A1A9ZH25 B4P0G1 A0A151J1J4 B4GX34 A0A1W4VWU8 A0A1B0B0S6 A0A1A9XD02 A0A0M4ECZ1 A0A232FBL4 A0A0M4ER64 A0A0M4E948 B4KEM1 B4M8N3 E1ZZV7 A0A0A1XT46 A0A0A1WX95
A0A2M4CGD1 W5JF67 A0A1Y9G9G1 A0A2M4BCK2 A0A2M4BCM7 A0A2M4BDM0 A0A182R3U4 A0A182Y0N9 A0A2M3Z1D4 A0A2M3ZE07 A0A182WGJ6 A0A182QWK5 A0A182K2I1 A0A182XN91 A0A182LZL0 A0A182JJ88 A0A182U8D7 A0A182UXG9 A0A240PL25 A0A182HRX0 Q7Q1J9 A0A182GI45 A0A1B6DUB7 A0A182GQM7 Q17HJ9 A0A182KU23 A0A1B6KA09 A0A1B6GNU4 A0A2J7QU96 A0A139WAY6 B0WR91 A0A1Y1M039 U4TXW0 A0A1J1IC50 A0A1B0GNY6 A0A084WU13 A0A2K9Y420 A0A2H8TCU4 A0A2S2PGF5 J9K345 A0A2S2PBL3 A0A2S2PXK3 A0A088AG98 A0A3Q0IXC2 A0A026VXF5 A0A3L8DDW7 E2CAC6 E0VYZ7 A0A158P0W6 A0A154PMJ3 A0A336N2A5 A0A336MZK1 A0A0C9RBF0 A0A0K8T5T2 A0A195CXY5 A0A1B0CG74 A0A0L7QVQ3 A0A310SFD5 A0A182SJM2 A0A023F3X6 K7IXA0 A0A151WW54 A0A0L0BM21 A0A1A9WU17 A0A1I8QEP7 Q8SXX2 B3MN04 B3NAA9 B4JCZ2 A0A3B0JNP4 B5DJZ6 B4HXC1 A0A0J9R277 A0A1A9ZH25 B4P0G1 A0A151J1J4 B4GX34 A0A1W4VWU8 A0A1B0B0S6 A0A1A9XD02 A0A0M4ECZ1 A0A232FBL4 A0A0M4ER64 A0A0M4E948 B4KEM1 B4M8N3 E1ZZV7 A0A0A1XT46 A0A0A1WX95
EC Number
3.4.-.-
Pubmed
22118469
26354079
20920257
23761445
25244985
12364791
+ More
26483478 17510324 20966253 18362917 19820115 28004739 23537049 24438588 24508170 30249741 20798317 20566863 21347285 26823975 25474469 20075255 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 15632085 22936249 17550304 28648823 18057021 25830018
26483478 17510324 20966253 18362917 19820115 28004739 23537049 24438588 24508170 30249741 20798317 20566863 21347285 26823975 25474469 20075255 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 15632085 22936249 17550304 28648823 18057021 25830018
EMBL
AGBW02010565
OWR48271.1
RSAL01000066
RVE49364.1
NWSH01006011
PCG63755.1
+ More
KQ459934 KPJ19244.1 KQ458714 KPJ05459.1 GGFL01000195 MBW64373.1 ADMH02001324 ETN63012.1 GGFJ01001638 MBW50779.1 GGFJ01001639 MBW50780.1 GGFJ01001966 MBW51107.1 GGFM01001578 MBW22329.1 GGFM01005960 MBW26711.1 AXCN02000684 AXCM01007372 APCN01001773 AAAB01008980 EAA14517.3 JXUM01065210 JXUM01065211 JXUM01065212 KQ562335 KXJ76121.1 GEDC01008045 JAS29253.1 JXUM01016019 KQ560447 KXJ82378.1 CH477248 EAT46108.1 GEBQ01031706 GEBQ01013511 JAT08271.1 JAT26466.1 GECZ01014580 GECZ01005674 JAS55189.1 JAS64095.1 NEVH01011186 PNF32150.1 KQ971380 KYB25031.1 DS232053 EDS33275.1 GEZM01044998 JAV77990.1 KB631815 ERL86439.1 CVRI01000044 CRK96558.1 AJVK01005614 ATLV01026973 ATLV01026974 ATLV01026975 ATLV01026976 ATLV01026977 ATLV01026978 ATLV01026979 ATLV01026980 ATLV01026981 ATLV01026982 KE525421 KFB53707.1 KY612467 AUW14597.1 GFXV01000118 MBW11923.1 GGMR01015669 MBY28288.1 ABLF02009599 ABLF02014513 GGMR01013687 MBY26306.1 GGMS01000887 MBY70090.1 KK107622 EZA48457.1 QOIP01000009 RLU18657.1 GL453975 EFN75092.1 DS235848 EEB18603.1 ADTU01005878 ADTU01005879 ADTU01005880 ADTU01005881 ADTU01005882 KQ434980 KZC13072.1 UFQT01002046 SSX32518.1 UFQS01003120 UFQT01003120 SSX15234.1 SSX34609.1 GBYB01013809 JAG83576.1 GBRD01005048 GBRD01005045 GDHC01019599 GDHC01011567 JAG60776.1 JAP99029.1 JAQ07062.1 KQ977141 KYN05442.1 AJWK01010804 AJWK01010805 KQ414726 KOC62616.1 KQ777390 OAD52110.1 GBBI01002586 JAC16126.1 AAZX01008641 AAZX01009269 KQ982691 KYQ52124.1 JRES01001649 KNC21170.1 AY075532 AE014134 AAL68339.1 ABC65898.1 CH902620 EDV31982.2 CH954177 EDV58611.2 CH916368 EDW03231.1 OUUW01000006 SPP82513.1 CH379061 EDY70621.1 CH480818 EDW51701.1 CM002910 KMY90188.1 CM000157 EDW88955.1 KQ980514 KYN15766.1 CH479195 EDW27361.1 JXJN01006810 JXJN01006811 CP012523 ALC39572.1 NNAY01000493 OXU28012.1 ALC39612.1 ALC39577.1 CH933807 EDW11900.1 KRG02965.1 CH940654 EDW57559.1 GL435530 EFN73268.1 GBXI01000177 JAD14115.1 GBXI01010820 JAD03472.1
KQ459934 KPJ19244.1 KQ458714 KPJ05459.1 GGFL01000195 MBW64373.1 ADMH02001324 ETN63012.1 GGFJ01001638 MBW50779.1 GGFJ01001639 MBW50780.1 GGFJ01001966 MBW51107.1 GGFM01001578 MBW22329.1 GGFM01005960 MBW26711.1 AXCN02000684 AXCM01007372 APCN01001773 AAAB01008980 EAA14517.3 JXUM01065210 JXUM01065211 JXUM01065212 KQ562335 KXJ76121.1 GEDC01008045 JAS29253.1 JXUM01016019 KQ560447 KXJ82378.1 CH477248 EAT46108.1 GEBQ01031706 GEBQ01013511 JAT08271.1 JAT26466.1 GECZ01014580 GECZ01005674 JAS55189.1 JAS64095.1 NEVH01011186 PNF32150.1 KQ971380 KYB25031.1 DS232053 EDS33275.1 GEZM01044998 JAV77990.1 KB631815 ERL86439.1 CVRI01000044 CRK96558.1 AJVK01005614 ATLV01026973 ATLV01026974 ATLV01026975 ATLV01026976 ATLV01026977 ATLV01026978 ATLV01026979 ATLV01026980 ATLV01026981 ATLV01026982 KE525421 KFB53707.1 KY612467 AUW14597.1 GFXV01000118 MBW11923.1 GGMR01015669 MBY28288.1 ABLF02009599 ABLF02014513 GGMR01013687 MBY26306.1 GGMS01000887 MBY70090.1 KK107622 EZA48457.1 QOIP01000009 RLU18657.1 GL453975 EFN75092.1 DS235848 EEB18603.1 ADTU01005878 ADTU01005879 ADTU01005880 ADTU01005881 ADTU01005882 KQ434980 KZC13072.1 UFQT01002046 SSX32518.1 UFQS01003120 UFQT01003120 SSX15234.1 SSX34609.1 GBYB01013809 JAG83576.1 GBRD01005048 GBRD01005045 GDHC01019599 GDHC01011567 JAG60776.1 JAP99029.1 JAQ07062.1 KQ977141 KYN05442.1 AJWK01010804 AJWK01010805 KQ414726 KOC62616.1 KQ777390 OAD52110.1 GBBI01002586 JAC16126.1 AAZX01008641 AAZX01009269 KQ982691 KYQ52124.1 JRES01001649 KNC21170.1 AY075532 AE014134 AAL68339.1 ABC65898.1 CH902620 EDV31982.2 CH954177 EDV58611.2 CH916368 EDW03231.1 OUUW01000006 SPP82513.1 CH379061 EDY70621.1 CH480818 EDW51701.1 CM002910 KMY90188.1 CM000157 EDW88955.1 KQ980514 KYN15766.1 CH479195 EDW27361.1 JXJN01006810 JXJN01006811 CP012523 ALC39572.1 NNAY01000493 OXU28012.1 ALC39612.1 ALC39577.1 CH933807 EDW11900.1 KRG02965.1 CH940654 EDW57559.1 GL435530 EFN73268.1 GBXI01000177 JAD14115.1 GBXI01010820 JAD03472.1
Proteomes
UP000007151
UP000283053
UP000218220
UP000053240
UP000053268
UP000075884
+ More
UP000000673 UP000069272 UP000075900 UP000076408 UP000075920 UP000075886 UP000075881 UP000076407 UP000075883 UP000075880 UP000075902 UP000075903 UP000075885 UP000075840 UP000007062 UP000069940 UP000249989 UP000008820 UP000075882 UP000235965 UP000007266 UP000002320 UP000030742 UP000183832 UP000092462 UP000030765 UP000007819 UP000005203 UP000079169 UP000053097 UP000279307 UP000008237 UP000009046 UP000005205 UP000076502 UP000078542 UP000092461 UP000053825 UP000075901 UP000002358 UP000075809 UP000037069 UP000091820 UP000095300 UP000000803 UP000007801 UP000008711 UP000001070 UP000268350 UP000001819 UP000001292 UP000092445 UP000002282 UP000078492 UP000008744 UP000192221 UP000092460 UP000092443 UP000092553 UP000215335 UP000009192 UP000008792 UP000000311
UP000000673 UP000069272 UP000075900 UP000076408 UP000075920 UP000075886 UP000075881 UP000076407 UP000075883 UP000075880 UP000075902 UP000075903 UP000075885 UP000075840 UP000007062 UP000069940 UP000249989 UP000008820 UP000075882 UP000235965 UP000007266 UP000002320 UP000030742 UP000183832 UP000092462 UP000030765 UP000007819 UP000005203 UP000079169 UP000053097 UP000279307 UP000008237 UP000009046 UP000005205 UP000076502 UP000078542 UP000092461 UP000053825 UP000075901 UP000002358 UP000075809 UP000037069 UP000091820 UP000095300 UP000000803 UP000007801 UP000008711 UP000001070 UP000268350 UP000001819 UP000001292 UP000092445 UP000002282 UP000078492 UP000008744 UP000192221 UP000092460 UP000092443 UP000092553 UP000215335 UP000009192 UP000008792 UP000000311
PRIDE
Interpro
SUPFAM
SSF49785
SSF49785
Gene 3D
CDD
ProteinModelPortal
A0A212F3F9
A0A3S2TLH5
A0A2A4IVE5
A0A0N1IC85
A0A194QJ89
A0A182NBR7
+ More
A0A2M4CGD1 W5JF67 A0A1Y9G9G1 A0A2M4BCK2 A0A2M4BCM7 A0A2M4BDM0 A0A182R3U4 A0A182Y0N9 A0A2M3Z1D4 A0A2M3ZE07 A0A182WGJ6 A0A182QWK5 A0A182K2I1 A0A182XN91 A0A182LZL0 A0A182JJ88 A0A182U8D7 A0A182UXG9 A0A240PL25 A0A182HRX0 Q7Q1J9 A0A182GI45 A0A1B6DUB7 A0A182GQM7 Q17HJ9 A0A182KU23 A0A1B6KA09 A0A1B6GNU4 A0A2J7QU96 A0A139WAY6 B0WR91 A0A1Y1M039 U4TXW0 A0A1J1IC50 A0A1B0GNY6 A0A084WU13 A0A2K9Y420 A0A2H8TCU4 A0A2S2PGF5 J9K345 A0A2S2PBL3 A0A2S2PXK3 A0A088AG98 A0A3Q0IXC2 A0A026VXF5 A0A3L8DDW7 E2CAC6 E0VYZ7 A0A158P0W6 A0A154PMJ3 A0A336N2A5 A0A336MZK1 A0A0C9RBF0 A0A0K8T5T2 A0A195CXY5 A0A1B0CG74 A0A0L7QVQ3 A0A310SFD5 A0A182SJM2 A0A023F3X6 K7IXA0 A0A151WW54 A0A0L0BM21 A0A1A9WU17 A0A1I8QEP7 Q8SXX2 B3MN04 B3NAA9 B4JCZ2 A0A3B0JNP4 B5DJZ6 B4HXC1 A0A0J9R277 A0A1A9ZH25 B4P0G1 A0A151J1J4 B4GX34 A0A1W4VWU8 A0A1B0B0S6 A0A1A9XD02 A0A0M4ECZ1 A0A232FBL4 A0A0M4ER64 A0A0M4E948 B4KEM1 B4M8N3 E1ZZV7 A0A0A1XT46 A0A0A1WX95
A0A2M4CGD1 W5JF67 A0A1Y9G9G1 A0A2M4BCK2 A0A2M4BCM7 A0A2M4BDM0 A0A182R3U4 A0A182Y0N9 A0A2M3Z1D4 A0A2M3ZE07 A0A182WGJ6 A0A182QWK5 A0A182K2I1 A0A182XN91 A0A182LZL0 A0A182JJ88 A0A182U8D7 A0A182UXG9 A0A240PL25 A0A182HRX0 Q7Q1J9 A0A182GI45 A0A1B6DUB7 A0A182GQM7 Q17HJ9 A0A182KU23 A0A1B6KA09 A0A1B6GNU4 A0A2J7QU96 A0A139WAY6 B0WR91 A0A1Y1M039 U4TXW0 A0A1J1IC50 A0A1B0GNY6 A0A084WU13 A0A2K9Y420 A0A2H8TCU4 A0A2S2PGF5 J9K345 A0A2S2PBL3 A0A2S2PXK3 A0A088AG98 A0A3Q0IXC2 A0A026VXF5 A0A3L8DDW7 E2CAC6 E0VYZ7 A0A158P0W6 A0A154PMJ3 A0A336N2A5 A0A336MZK1 A0A0C9RBF0 A0A0K8T5T2 A0A195CXY5 A0A1B0CG74 A0A0L7QVQ3 A0A310SFD5 A0A182SJM2 A0A023F3X6 K7IXA0 A0A151WW54 A0A0L0BM21 A0A1A9WU17 A0A1I8QEP7 Q8SXX2 B3MN04 B3NAA9 B4JCZ2 A0A3B0JNP4 B5DJZ6 B4HXC1 A0A0J9R277 A0A1A9ZH25 B4P0G1 A0A151J1J4 B4GX34 A0A1W4VWU8 A0A1B0B0S6 A0A1A9XD02 A0A0M4ECZ1 A0A232FBL4 A0A0M4ER64 A0A0M4E948 B4KEM1 B4M8N3 E1ZZV7 A0A0A1XT46 A0A0A1WX95
PDB
5A2R
E-value=4.48679e-122,
Score=1123
Ontologies
GO
PANTHER
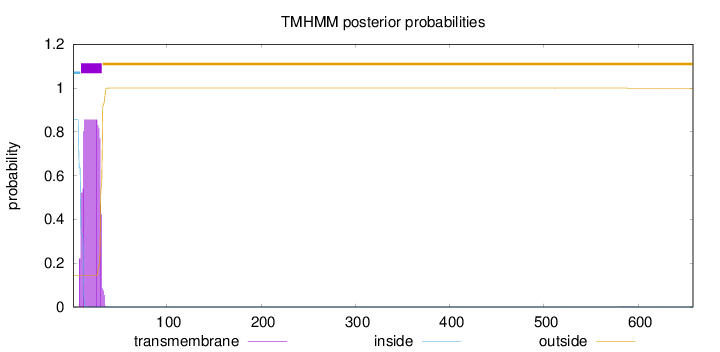
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.764362
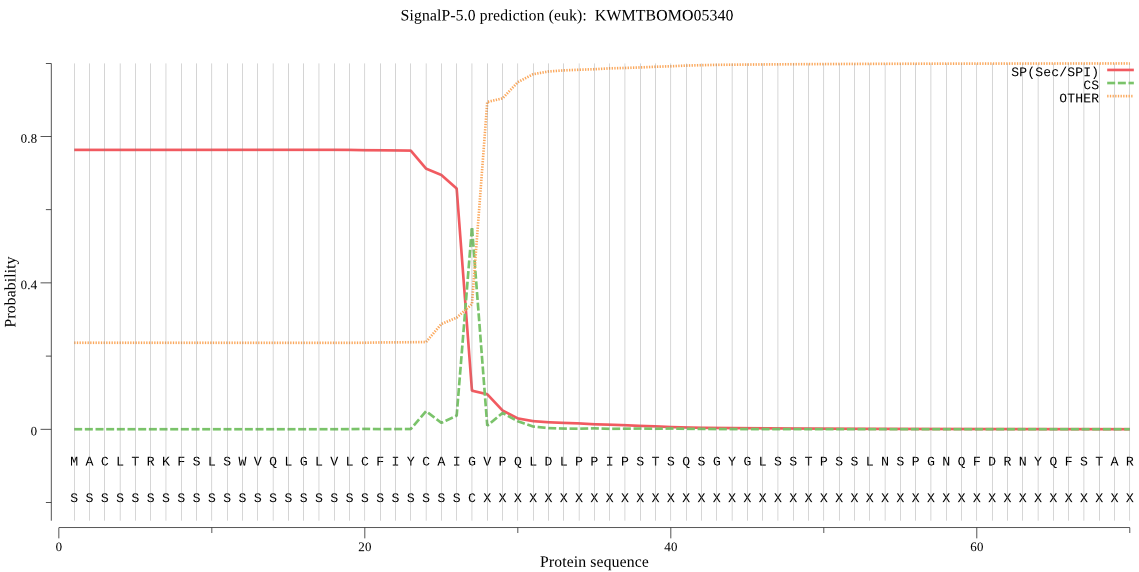
Length:
658
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.34941
Exp number, first 60 AAs:
18.34409
Total prob of N-in:
0.85621
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 658
Population Genetic Test Statistics
Pi
202.740497
Theta
154.842638
Tajima's D
1.125536
CLR
0.633938
CSRT
0.696815159242038
Interpretation
Uncertain
