Gene
KWMTBOMO05339 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002527
Annotation
male_reproductive_organ_angiotensin-converting_enzyme-related_protein_2_precursor_[Bombyx_mori]
Full name
Angiotensin-converting enzyme
Location in the cell
Cytoplasmic Reliability : 3.183
Sequence
CDS
ATGATAATGAAGATTATCGGGGAGCTCGCCCTTCTGGGTGCTATCGCGTCAGTCTTCCTGGTAGCAGCCCAGGGTCGGGCAGCAGAAAGAGCTGCAGAGGAATCCGCTGGGCGAGATTACGTGAGGACTTTGGACGAACAGACTTCTCGGCTCAGGAACAAGAAAACGATTGCCTCTTGGGACTATGAGACGAATTTGACAGTCCATAACGAACAGAAATTGTTACAAGTATCCTTAGAGGTGGCAAGCGAAACGAAGGAATACTGGAAGGAAACCATGAGCTACCTCTGGCACGAGTTCGAAGACCCGGACCTGAAGAGGATGTTCAAGAAATACACGGAGCTGGGCACATCAGCTCTACCCGAGGATTTGAATCAGAGATTGATCGTGGCCATCAATAGCATGCAGTCAACTTACGCTAAGGCAACGATCTGTGATTATAATAATCGGACTAAATGTGATTTGCATGTTGAACCTGAAATCACAAACATTTTTGCGACAAGCAGCGACGCTGAAGAACTCAAGTACACATGGCTTGAATGGCATAAGGCAGCGGGTGCTCCATCGAAGAGTAACTTCACCGAGTACGTTCAAATGGAGAACGAGGCTGCCAAATTAAATGGTTACGACACTGTGGCCGAATGGTGGCTTAGTGAATATGAAGCTACAGACAGCGAGGACCAGTTCGCAGCGCTCTGGGCGCAAATCAAACCGCTCTATGAACAGATTCATGCGTACGTCAGAGGGCATCTTCGTGTGAAGTATGGAGAAGAAGTCGTGCCGAAGAAAGGACCCATTCCAGCTCATTTGTTAGGTAACATTTGGGCCCAAACTTGGGGTAACGTTGAAAAATTCACAAGGCCGTATCCGGATAAGACCGATATTGATGTAACTGCCGCGCTCATTGCACAGAATTACACAGCCCTTAAGATGTTTAAGACAGCAGAGGAGTTTTTTACGTCATTGGATTTATCCCCAATGCCAGAGTTGTTCTGGGAAAGATCCATCTTAGAGAAACCAACTGATGGAAGAGAAATGGTCTGCCATGCTTCTGCTTGGGACTTTTTCGACGGTGAAGATTTCAGAATCCGTCAATGCACAACCATCACGGACGCGTTCTTCAAGACGACCCACCACGAAATGGGTCACATACAATACTACTTGCAGTACAAGGATCAGCCGGTGATCTACAGGGCTGGAGCTAATCCAGGTTTTCACGAAGCAGTTGGTGACGTCATCGCACTGTCGGTGGCAACGCCGAAACATTTACGGGTGATGGGGCTTCTCGAGGATGGACCAGAAGATTTGGAATCTAACATTAATCAGCTTTACAAAATGGGTCTGGACAAGATAGTTTTCTTACCATTCGGCTATCTCCTCGATCTCTTCCGTTATGGCGTCTTCCGAGGAACTACCACAGTTGACGACTACAATTGTCACTTCTGGCAACTAAGAGAGTCCTTGCAGGGCGTGGAACCTCCGGCGCCACGAAGCGAAGCGGACTTTGATCCTGCTGCAAAGTACCATATAGCTGCAGATGTGGAATACATGAGATACTATATCTCTTACATTATCCAGTTTCAATTCCATCGTTCGTTATGCCAGCTTGCCGGGGAGTACGCGGTTGGCGACTCCAGCAGGCTGCTCTCAAACTGCGATATTTACAACAGCACGGCGGCAGGAAACGCTCTCGGCAAAATGCTGCAATTGGGTTCTTCGAAGCCGTGGCCCGACGCGATGGAAGCGATAACTGGACAAAGGTTCATGGATGCCAGCGGCGTCCTGGAGTACTTCCAGCCTCTATACGATTGGCTCAAGGCCGAGAACGAACGAAATGGAGAATTTGTCGGTTGGGAGCCTAGTACCGTCCAATTTTGTACTCCAAACCAAAGAGCAGCGATGGAAGCGACCGGCTTTAACAAGAGATTGAAGAAATATGCAGCTCTATAG
Protein
MIMKIIGELALLGAIASVFLVAAQGRAAERAAEESAGRDYVRTLDEQTSRLRNKKTIASWDYETNLTVHNEQKLLQVSLEVASETKEYWKETMSYLWHEFEDPDLKRMFKKYTELGTSALPEDLNQRLIVAINSMQSTYAKATICDYNNRTKCDLHVEPEITNIFATSSDAEELKYTWLEWHKAAGAPSKSNFTEYVQMENEAAKLNGYDTVAEWWLSEYEATDSEDQFAALWAQIKPLYEQIHAYVRGHLRVKYGEEVVPKKGPIPAHLLGNIWAQTWGNVEKFTRPYPDKTDIDVTAALIAQNYTALKMFKTAEEFFTSLDLSPMPELFWERSILEKPTDGREMVCHASAWDFFDGEDFRIRQCTTITDAFFKTTHHEMGHIQYYLQYKDQPVIYRAGANPGFHEAVGDVIALSVATPKHLRVMGLLEDGPEDLESNINQLYKMGLDKIVFLPFGYLLDLFRYGVFRGTTTVDDYNCHFWQLRESLQGVEPPAPRSEADFDPAAKYHIAADVEYMRYYISYIIQFQFHRSLCQLAGEYAVGDSSRLLSNCDIYNSTAAGNALGKMLQLGSSKPWPDAMEAITGQRFMDASGVLEYFQPLYDWLKAENERNGEFVGWEPSTVQFCTPNQRAAMEATGFNKRLKKYAAL
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M2 family.
Feature
chain Angiotensin-converting enzyme
Uniprot
H9IZ42
B9X255
B0LJE2
A0A2H1VQG9
A0A212F3H8
A0A3S2M2A7
+ More
A0A1E1W694 A0A0N0PEJ7 A0A2A4IWT8 A0A0L7LS53 H9IZ41 Q9NDS8 A0A212F3I4 A0A194QIX2 A0A3S2LLM4 A0A0N1ICA6 A0A2H1V7I0 I4DJQ5 E0VYZ8 A0A0U3KDL0 A0A1Q3FRW0 A0A2J7QUF6 A0A2J7QUE9 A0A1S4G6D0 J9E9A1 A0A1Y1NHL6 A0A1Q3FRM3 Q6RX62 A0A0N0BEI9 A0A1I9WL53 A0A023EUI3 A0A023EVK4 A0A023EVB9 A0A023EXC8 A0A087ZRC1 A0A310SP14 M9PM20 B0WR90 W5JI94 V9IK38 A0A2A3E9I4 A0A026X3N5 D6X4L0 A0A182GI44 A0A1S6J0Y7 A0A2M4AK25 A0A2M4AJS0 A0A2M4AK82 A0A2M4AJQ6 A0A2M4DP36 T1DLB0 A0A182XN90 A0A1Y9G9N3 A0A0L7QKN3 A0A2J7QUG0 A0NFV1 A0A2C9GR30 A0A2M4BH35 A0A2M4BH75 A0A2M4BH14 A0A182Y0N8 A0A2M4BGX1 A0A2M4BHR7 A0A182LUT3 A0A1Y9G8A7 A0A182QDG1 A0A154PK20 A0A084VR60 A0A1I8JUB7 A0A182GQM8 A0A1Y9G8E7 A0A084VR61 A0A240PM54 A0A182WGJ2 Q7PM22 A0A1Q3FFN8 A0A1Q3FFN2 A0A1B6EFN8 A0NFV2 A0A240PPV9 A0A182JL55 A0A1Q3FG20 A0A182NBR7 A0A1B6C745 A0A2C9GRB4 A0A1B6DC58 A0A1Q3FG39 Q5WPT4 A0A1L8DR16 A0A182QUK4 A0A2S2QGZ9 T1EB36 A0A182J606 A0A336LZ65 J9K7M4 A0A182Y0N9 A0A182TYM7 A0A1Y9G9F7 A0A1I8JUD0 A0A182KU25
A0A1E1W694 A0A0N0PEJ7 A0A2A4IWT8 A0A0L7LS53 H9IZ41 Q9NDS8 A0A212F3I4 A0A194QIX2 A0A3S2LLM4 A0A0N1ICA6 A0A2H1V7I0 I4DJQ5 E0VYZ8 A0A0U3KDL0 A0A1Q3FRW0 A0A2J7QUF6 A0A2J7QUE9 A0A1S4G6D0 J9E9A1 A0A1Y1NHL6 A0A1Q3FRM3 Q6RX62 A0A0N0BEI9 A0A1I9WL53 A0A023EUI3 A0A023EVK4 A0A023EVB9 A0A023EXC8 A0A087ZRC1 A0A310SP14 M9PM20 B0WR90 W5JI94 V9IK38 A0A2A3E9I4 A0A026X3N5 D6X4L0 A0A182GI44 A0A1S6J0Y7 A0A2M4AK25 A0A2M4AJS0 A0A2M4AK82 A0A2M4AJQ6 A0A2M4DP36 T1DLB0 A0A182XN90 A0A1Y9G9N3 A0A0L7QKN3 A0A2J7QUG0 A0NFV1 A0A2C9GR30 A0A2M4BH35 A0A2M4BH75 A0A2M4BH14 A0A182Y0N8 A0A2M4BGX1 A0A2M4BHR7 A0A182LUT3 A0A1Y9G8A7 A0A182QDG1 A0A154PK20 A0A084VR60 A0A1I8JUB7 A0A182GQM8 A0A1Y9G8E7 A0A084VR61 A0A240PM54 A0A182WGJ2 Q7PM22 A0A1Q3FFN8 A0A1Q3FFN2 A0A1B6EFN8 A0NFV2 A0A240PPV9 A0A182JL55 A0A1Q3FG20 A0A182NBR7 A0A1B6C745 A0A2C9GRB4 A0A1B6DC58 A0A1Q3FG39 Q5WPT4 A0A1L8DR16 A0A182QUK4 A0A2S2QGZ9 T1EB36 A0A182J606 A0A336LZ65 J9K7M4 A0A182Y0N9 A0A182TYM7 A0A1Y9G9F7 A0A1I8JUD0 A0A182KU25
EC Number
3.4.-.-
Pubmed
EMBL
BABH01011115
BABH01011116
AB485781
BAH23569.1
EU170161
ABW34729.1
+ More
ODYU01003823 SOQ43060.1 AGBW02010565 OWR48272.1 RSAL01000066 RVE49365.1 GDQN01008554 JAT82500.1 KQ459934 KPJ19243.1 NWSH01006011 PCG63754.1 JTDY01000210 KOB78277.1 AB026110 AB485780 BAA97657.1 BAH23568.1 OWR48273.1 KQ458714 KPJ05462.1 RVE49366.1 KQ461002 KPJ10041.1 ODYU01000895 SOQ36342.1 AK401523 BAM18145.1 DS235848 EEB18604.1 KU218691 ALX00071.1 GFDL01004760 JAV30285.1 NEVH01011186 PNF32210.1 PNF32208.1 CH477248 EJY57431.1 GEZM01002421 JAV97269.1 GFDL01004795 JAV30250.1 AY487174 AAR85358.1 KQ435827 KOX71968.1 KU932237 APA33873.1 GAPW01000528 JAC13070.1 GAPW01000531 JAC13067.1 GAPW01000530 JAC13068.1 GAPW01000529 JAC13069.1 KQ762196 OAD56126.1 KC437298 AGC79111.1 DS232053 EDS33274.1 ADMH02001324 ETN63008.1 JR048421 AEY60659.1 KZ288322 PBC28134.1 KK107013 QOIP01000003 EZA62925.1 RLU25153.1 KQ971380 EEZ97252.2 JXUM01065208 JXUM01065209 KQ562335 KXJ76120.1 KU764440 AQS60686.1 GGFK01007818 MBW41139.1 GGFK01007715 MBW41036.1 GGFK01007707 MBW41028.1 GGFK01007699 MBW41020.1 GGFL01015113 MBW79291.1 GAMD01000546 JAB01045.1 KQ414954 KOC59071.1 PNF32211.1 AAAB01008980 EAU76040.2 APCN01001774 GGFJ01003206 MBW52347.1 GGFJ01003160 MBW52301.1 GGFJ01003208 MBW52349.1 GGFJ01003159 MBW52300.1 GGFJ01003207 MBW52348.1 AXCM01007217 AXCN02000684 KQ434943 KZC12209.1 ATLV01015459 KE525014 KFB40454.1 JXUM01016020 JXUM01016021 KQ560447 KXJ82379.1 KFB40455.1 EAA14501.5 GFDL01008681 JAV26364.1 GFDL01008678 JAV26367.1 GEDC01000545 JAS36753.1 EAU76041.1 GFDL01008546 JAV26499.1 GEDC01028238 JAS09060.1 GEDC01024908 GEDC01014015 JAS12390.1 JAS23283.1 GFDL01008526 JAV26519.1 AY455911 AAS16911.1 GFDF01005267 JAV08817.1 GGMS01007793 MBY76996.1 GAMD01000383 JAB01208.1 UFQS01000326 UFQT01000326 SSX02915.1 SSX23282.1 ABLF02025366
ODYU01003823 SOQ43060.1 AGBW02010565 OWR48272.1 RSAL01000066 RVE49365.1 GDQN01008554 JAT82500.1 KQ459934 KPJ19243.1 NWSH01006011 PCG63754.1 JTDY01000210 KOB78277.1 AB026110 AB485780 BAA97657.1 BAH23568.1 OWR48273.1 KQ458714 KPJ05462.1 RVE49366.1 KQ461002 KPJ10041.1 ODYU01000895 SOQ36342.1 AK401523 BAM18145.1 DS235848 EEB18604.1 KU218691 ALX00071.1 GFDL01004760 JAV30285.1 NEVH01011186 PNF32210.1 PNF32208.1 CH477248 EJY57431.1 GEZM01002421 JAV97269.1 GFDL01004795 JAV30250.1 AY487174 AAR85358.1 KQ435827 KOX71968.1 KU932237 APA33873.1 GAPW01000528 JAC13070.1 GAPW01000531 JAC13067.1 GAPW01000530 JAC13068.1 GAPW01000529 JAC13069.1 KQ762196 OAD56126.1 KC437298 AGC79111.1 DS232053 EDS33274.1 ADMH02001324 ETN63008.1 JR048421 AEY60659.1 KZ288322 PBC28134.1 KK107013 QOIP01000003 EZA62925.1 RLU25153.1 KQ971380 EEZ97252.2 JXUM01065208 JXUM01065209 KQ562335 KXJ76120.1 KU764440 AQS60686.1 GGFK01007818 MBW41139.1 GGFK01007715 MBW41036.1 GGFK01007707 MBW41028.1 GGFK01007699 MBW41020.1 GGFL01015113 MBW79291.1 GAMD01000546 JAB01045.1 KQ414954 KOC59071.1 PNF32211.1 AAAB01008980 EAU76040.2 APCN01001774 GGFJ01003206 MBW52347.1 GGFJ01003160 MBW52301.1 GGFJ01003208 MBW52349.1 GGFJ01003159 MBW52300.1 GGFJ01003207 MBW52348.1 AXCM01007217 AXCN02000684 KQ434943 KZC12209.1 ATLV01015459 KE525014 KFB40454.1 JXUM01016020 JXUM01016021 KQ560447 KXJ82379.1 KFB40455.1 EAA14501.5 GFDL01008681 JAV26364.1 GFDL01008678 JAV26367.1 GEDC01000545 JAS36753.1 EAU76041.1 GFDL01008546 JAV26499.1 GEDC01028238 JAS09060.1 GEDC01024908 GEDC01014015 JAS12390.1 JAS23283.1 GFDL01008526 JAV26519.1 AY455911 AAS16911.1 GFDF01005267 JAV08817.1 GGMS01007793 MBY76996.1 GAMD01000383 JAB01208.1 UFQS01000326 UFQT01000326 SSX02915.1 SSX23282.1 ABLF02025366
Proteomes
UP000005204
UP000007151
UP000283053
UP000053240
UP000218220
UP000037510
+ More
UP000053268 UP000009046 UP000235965 UP000008820 UP000053105 UP000005203 UP000002320 UP000000673 UP000242457 UP000053097 UP000279307 UP000007266 UP000069940 UP000249989 UP000076407 UP000069272 UP000053825 UP000007062 UP000075840 UP000076408 UP000075883 UP000075886 UP000076502 UP000030765 UP000075900 UP000075885 UP000075920 UP000075880 UP000075884 UP000007819 UP000075902 UP000075882
UP000053268 UP000009046 UP000235965 UP000008820 UP000053105 UP000005203 UP000002320 UP000000673 UP000242457 UP000053097 UP000279307 UP000007266 UP000069940 UP000249989 UP000076407 UP000069272 UP000053825 UP000007062 UP000075840 UP000076408 UP000075883 UP000075886 UP000076502 UP000030765 UP000075900 UP000075885 UP000075920 UP000075880 UP000075884 UP000007819 UP000075902 UP000075882
Gene 3D
CDD
ProteinModelPortal
H9IZ42
B9X255
B0LJE2
A0A2H1VQG9
A0A212F3H8
A0A3S2M2A7
+ More
A0A1E1W694 A0A0N0PEJ7 A0A2A4IWT8 A0A0L7LS53 H9IZ41 Q9NDS8 A0A212F3I4 A0A194QIX2 A0A3S2LLM4 A0A0N1ICA6 A0A2H1V7I0 I4DJQ5 E0VYZ8 A0A0U3KDL0 A0A1Q3FRW0 A0A2J7QUF6 A0A2J7QUE9 A0A1S4G6D0 J9E9A1 A0A1Y1NHL6 A0A1Q3FRM3 Q6RX62 A0A0N0BEI9 A0A1I9WL53 A0A023EUI3 A0A023EVK4 A0A023EVB9 A0A023EXC8 A0A087ZRC1 A0A310SP14 M9PM20 B0WR90 W5JI94 V9IK38 A0A2A3E9I4 A0A026X3N5 D6X4L0 A0A182GI44 A0A1S6J0Y7 A0A2M4AK25 A0A2M4AJS0 A0A2M4AK82 A0A2M4AJQ6 A0A2M4DP36 T1DLB0 A0A182XN90 A0A1Y9G9N3 A0A0L7QKN3 A0A2J7QUG0 A0NFV1 A0A2C9GR30 A0A2M4BH35 A0A2M4BH75 A0A2M4BH14 A0A182Y0N8 A0A2M4BGX1 A0A2M4BHR7 A0A182LUT3 A0A1Y9G8A7 A0A182QDG1 A0A154PK20 A0A084VR60 A0A1I8JUB7 A0A182GQM8 A0A1Y9G8E7 A0A084VR61 A0A240PM54 A0A182WGJ2 Q7PM22 A0A1Q3FFN8 A0A1Q3FFN2 A0A1B6EFN8 A0NFV2 A0A240PPV9 A0A182JL55 A0A1Q3FG20 A0A182NBR7 A0A1B6C745 A0A2C9GRB4 A0A1B6DC58 A0A1Q3FG39 Q5WPT4 A0A1L8DR16 A0A182QUK4 A0A2S2QGZ9 T1EB36 A0A182J606 A0A336LZ65 J9K7M4 A0A182Y0N9 A0A182TYM7 A0A1Y9G9F7 A0A1I8JUD0 A0A182KU25
A0A1E1W694 A0A0N0PEJ7 A0A2A4IWT8 A0A0L7LS53 H9IZ41 Q9NDS8 A0A212F3I4 A0A194QIX2 A0A3S2LLM4 A0A0N1ICA6 A0A2H1V7I0 I4DJQ5 E0VYZ8 A0A0U3KDL0 A0A1Q3FRW0 A0A2J7QUF6 A0A2J7QUE9 A0A1S4G6D0 J9E9A1 A0A1Y1NHL6 A0A1Q3FRM3 Q6RX62 A0A0N0BEI9 A0A1I9WL53 A0A023EUI3 A0A023EVK4 A0A023EVB9 A0A023EXC8 A0A087ZRC1 A0A310SP14 M9PM20 B0WR90 W5JI94 V9IK38 A0A2A3E9I4 A0A026X3N5 D6X4L0 A0A182GI44 A0A1S6J0Y7 A0A2M4AK25 A0A2M4AJS0 A0A2M4AK82 A0A2M4AJQ6 A0A2M4DP36 T1DLB0 A0A182XN90 A0A1Y9G9N3 A0A0L7QKN3 A0A2J7QUG0 A0NFV1 A0A2C9GR30 A0A2M4BH35 A0A2M4BH75 A0A2M4BH14 A0A182Y0N8 A0A2M4BGX1 A0A2M4BHR7 A0A182LUT3 A0A1Y9G8A7 A0A182QDG1 A0A154PK20 A0A084VR60 A0A1I8JUB7 A0A182GQM8 A0A1Y9G8E7 A0A084VR61 A0A240PM54 A0A182WGJ2 Q7PM22 A0A1Q3FFN8 A0A1Q3FFN2 A0A1B6EFN8 A0NFV2 A0A240PPV9 A0A182JL55 A0A1Q3FG20 A0A182NBR7 A0A1B6C745 A0A2C9GRB4 A0A1B6DC58 A0A1Q3FG39 Q5WPT4 A0A1L8DR16 A0A182QUK4 A0A2S2QGZ9 T1EB36 A0A182J606 A0A336LZ65 J9K7M4 A0A182Y0N9 A0A182TYM7 A0A1Y9G9F7 A0A1I8JUD0 A0A182KU25
PDB
5A2R
E-value=0,
Score=1632
Ontologies
KEGG
GO
PANTHER
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.602732
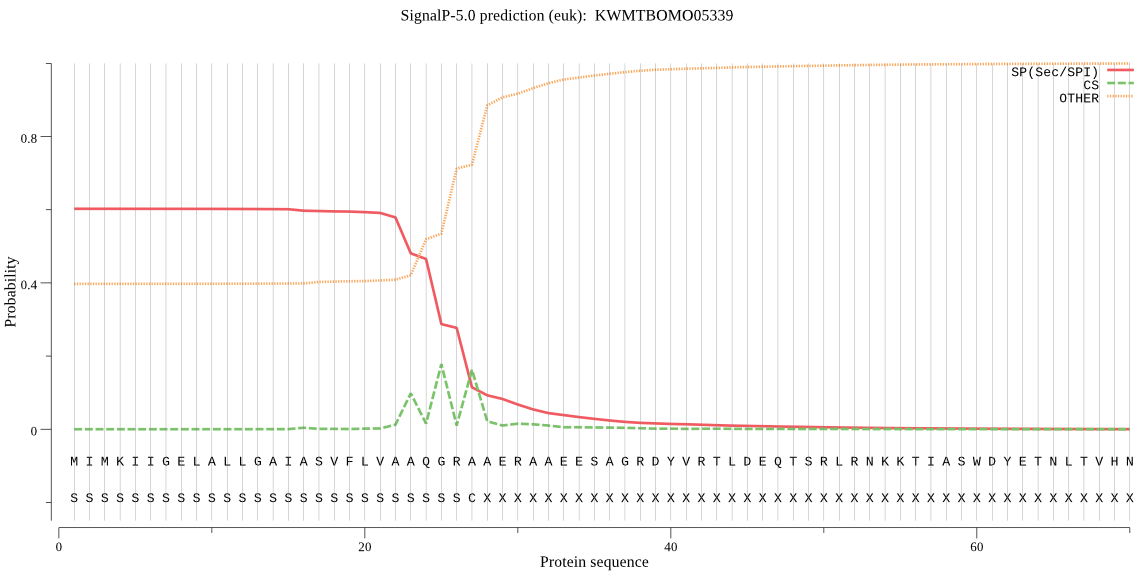
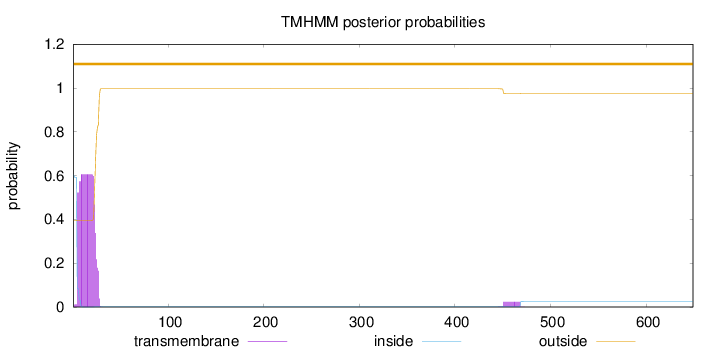
Length:
649
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.55305
Exp number, first 60 AAs:
12.10793
Total prob of N-in:
0.60419
POSSIBLE N-term signal
sequence
outside
1 - 649
Population Genetic Test Statistics
Pi
269.074152
Theta
137.055457
Tajima's D
2.988655
CLR
0.313824
CSRT
0.980800959952002
Interpretation
Uncertain
