Gene
KWMTBOMO05333
Pre Gene Modal
BGIBMGA002389
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.285
Sequence
CDS
ATGGCTGGAAGTTACATTGTCAACGTGCCTAAATTGAAAGGTCGCGAAAACTTCGACGAGTGGGCGTTTGCAGCTGAACATTTTTTGATGTTGGAAGGTGTCGATATTCATAAGTTAGAATCATCAACCGATGCTAGTTCTATTGATGAAAGAAAAGCTAAAGCTAAACTTATTATGACAATTGACTCTTCAATTTATGTTCACATTAAAAACGAGCAAACAGTACAAGGTGTGTGGAAGCGTTTAAAAGCCCTGTACGATGACTGTGGCTTTACGCGTCGCATCAGTCTACTTCGTCAGCTAATATCAATTCGATTAGAAGATAGCGAATCAATGGAAGCATATGTAACGCAAATGGTCGACACTGCACAAAAACTTAAAGGTACGGGTTTTCAGATCACTGATGAGTGGATCGGTTCACTATTATTAGCGGGCCTAACAGATACATTTGCTCCTATGATAATGGCCATTGAACATTCTGGATTAGCCATCAGTACGGATGCCATCAAGTCAAAATTACTGGACATGAGTTCAGAAGTTGGCAACACTGCAACTAACAATGCCTTTTTGTCGAAAGTAAGTCAGATGATTCGTAATGGAAACAAAGTGAGCTTCGAAGAAGACGTTTGTTACATACGCAATCGACAGAATGTGCTGGTAGGAAAAGCAAACCTGGTGAATGGAGTCTATAAATTGGACATTAAGTCAGACAGTTTGTTCGCGTGCTCAGCCACAACTGTGACTAGTGAGATATGGCATCGAAGATTAGGACACATTAACAGTAGAAGCTTGAATATTATGAAGAATGGTGCTGTGGATGGGATATCATACATGGATAAAGCTGAAATTGATATGTCCAAGTGCATCATATGCTGTGAAGGAAAACAGTCAAGGTTGCCATTTTCACATAAAGGTACGAGAAGTGAAAATGTGCTTGAACTTGTTCATGCAGATGTCTGTGGACCCATGGAACAACCTTCTATTGGACTATCAAAGTATTTTTTGCTTTTTGTAGATGACTACAGTAGAATGGCATTTGTGTTTTTTCTCAAAGCCAAGAACCGAGTTTTCAAATATTTTAAAGAGTTCAAAACTCTGGTTGAGACACAGACAGGCAGAAAACTAAAAGCCTTAAGGACAGATAATGGTGGAGAGTTTTGTTCGCTAGAATTTGATGAGTTTTTGAAGAAAAATGGCATAGTACATCAAACAACAAACCCTCATACACCAGAACAGAATGGCCTGTGTGAAAGACTTAATAGAACTGTCGTGGAGAGAGCTCGGTGTCTTCTGTTTGAAGCAGGCCTAGATAAAAAGTTTTGGGCCGAAGCAATCAACACAGCAGTCTATTTACGAAATAGAACATTGGCAGCAGGACTTAGGAAAACACCTTATGAAATATGGAGTGGTAAAAAGCCAAATCTAAGTCATATACGGATATTTGGTAGTCCTGCAATGGTCCATGTCCCCAAAGCAAAGCGGACTAAATGGAACAAGAAGGCCAATGAACACATCCTGATTGGATACTCAGAGAGTACCAAAGGATACAGGCTGTACAATGTAAAAACAAAAAATATTACCACTAGCAGGGATGTTGTTATCATGGAGAAAATCAACGACACAGATATGATACAAGCAGTAGTGGAAGAAAAAGAAACTATTCCAAGTGAAGAAGGAATACAAAAATGTGCTGTGACAAGTGAAGATTCCACTTGTGACTCACTAAATAGCACATACATGACAGATGATGCTACATATGTACCAGATGATACCAGTACCACCAGCTCAGATGAATTCTGTGATAGTAAATCGAATCACAAGTCAGAAAATCTGCAGCCAATAATTAAGGTGGAAAAGAGGACTAGGAAGAAACCAGACTGGTATGGGTACAATAACATATGTCAGAGCAGTGAGGAGGTGGACGACAATCTTCTCACTTATGAGGATGTAATGAGAGGATCAGAACAAGAAGAATGGTCAAAAGCCATGAAAGAGGAAATACAATCTTTCCATCAGAATGATGCTTGGGAGTTAGTAGACAGACCCAGTGGTGAAAATATTGTGGCATGCAAATGGGTGTATAAAAAGAAACTTAACAGTGACAACTCAGTGAGATATAGGGCACGGTTGGTTGCGAAGGGTTTCAGCCAGAGACAGGGTATAGACTTTAAAGAAACTTTTTCCCCAGTGCTAAAATATTCCACTTTAAAACTGTTATTTGCTATTAGTGTTAACTTGAATCTTAGCATAACCCATTTGGATGTAACGACGGCCTTTTTGAATGGTCACCTAGATGAAACTGTTTTTATGCAATTGCCTCAGAGTTTTGAATGTAAACCTAATGCTGATAAAGTCTTGAAATTGAAAAAGGCCATTTATGGACTAAAACAGTCAGCGAGGGCCTGGCTGTTTAGTTTCTCATGA
Protein
MAGSYIVNVPKLKGRENFDEWAFAAEHFLMLEGVDIHKLESSTDASSIDERKAKAKLIMTIDSSIYVHIKNEQTVQGVWKRLKALYDDCGFTRRISLLRQLISIRLEDSESMEAYVTQMVDTAQKLKGTGFQITDEWIGSLLLAGLTDTFAPMIMAIEHSGLAISTDAIKSKLLDMSSEVGNTATNNAFLSKVSQMIRNGNKVSFEEDVCYIRNRQNVLVGKANLVNGVYKLDIKSDSLFACSATTVTSEIWHRRLGHINSRSLNIMKNGAVDGISYMDKAEIDMSKCIICCEGKQSRLPFSHKGTRSENVLELVHADVCGPMEQPSIGLSKYFLLFVDDYSRMAFVFFLKAKNRVFKYFKEFKTLVETQTGRKLKALRTDNGGEFCSLEFDEFLKKNGIVHQTTNPHTPEQNGLCERLNRTVVERARCLLFEAGLDKKFWAEAINTAVYLRNRTLAAGLRKTPYEIWSGKKPNLSHIRIFGSPAMVHVPKAKRTKWNKKANEHILIGYSESTKGYRLYNVKTKNITTSRDVVIMEKINDTDMIQAVVEEKETIPSEEGIQKCAVTSEDSTCDSLNSTYMTDDATYVPDDTSTTSSDEFCDSKSNHKSENLQPIIKVEKRTRKKPDWYGYNNICQSSEEVDDNLLTYEDVMRGSEQEEWSKAMKEEIQSFHQNDAWELVDRPSGENIVACKWVYKKKLNSDNSVRYRARLVAKGFSQRQGIDFKETFSPVLKYSTLKLLFAISVNLNLSITHLDVTTAFLNGHLDETVFMQLPQSFECKPNADKVLKLKKAIYGLKQSARAWLFSFS
Summary
Uniprot
A0A1Y1N668
A0A1Y1NAZ7
A0A1Y1N664
A0A1Y1N932
A0A0V1APU2
A0A146LH62
+ More
A0A355BEE2 A0A0A9XY84 A0A194RJV9 A0A0A1WXQ3 A0A034VP70 A0A194RU38 A0A0J7NHJ8 A0A2A4JRI1 A0A2J7Q1Z2 A0A2J7R668 A0A2J7RKJ6 A0A2J7R8S8 A0A0N1ICY1 A0A1Y3EBW8 A0A0V0T3I0 A0A2A4J959 A0A2J7R8R7 A0A0V0TUP0 A0A1Y1N3X2 A0A085NU43 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A0V1F2E6 A0A085MVP5 A0A0V0YNF4 A0A1Y1LYU4 A0A085N572 A0A1Y1LP39 A0A251T2S6 A0A1W7R6K6 A0A1W7R6J6 A0A1X7U495 A0A0A1WFC7 T1PKV2 A0A251TN47 A0A0J7KG03 A0A251TCA0 A0A251T6N3 A0A0A9WPH5 A0A0K8U6S2 A0A2B4R8Q4 W8AEF0 A0A392M204 A0A0K8U8C3 A0A0V0IV83 A0A151RZS8 A0A085ND65 A0A251UIF2 A0A224XA26 Q6L3N8 A0A0A1XLZ7 W8AQ86 W8AL23 A0A0V0Z6Q0 A0A0A9WRE3 A0A396INR0 A0A0K8U6X2 A0A0A9YB53 A0A0A9W6P7 W8B7K5 A0A0K8WJW9 A0A151T5R0 A0A151TGQ2 A0A2J7PSB2 W8BYI7 A0A1W7R6A5 A0A151THM1 A0A2N9ETM3 A0A0J7K6W1 A0A2N9GX95 A0A2N9IWX1 A0A2N9GK87 Q5MG99 A0A2K3MYT4 A0A151R2G6 A0A2N9HR09 A0A151TGP0 A0A2N9ELX4 A0A2N9IUL9 A0A0V0RWJ3 Q7Y141 A0A151TPU3
A0A355BEE2 A0A0A9XY84 A0A194RJV9 A0A0A1WXQ3 A0A034VP70 A0A194RU38 A0A0J7NHJ8 A0A2A4JRI1 A0A2J7Q1Z2 A0A2J7R668 A0A2J7RKJ6 A0A2J7R8S8 A0A0N1ICY1 A0A1Y3EBW8 A0A0V0T3I0 A0A2A4J959 A0A2J7R8R7 A0A0V0TUP0 A0A1Y1N3X2 A0A085NU43 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A0V1F2E6 A0A085MVP5 A0A0V0YNF4 A0A1Y1LYU4 A0A085N572 A0A1Y1LP39 A0A251T2S6 A0A1W7R6K6 A0A1W7R6J6 A0A1X7U495 A0A0A1WFC7 T1PKV2 A0A251TN47 A0A0J7KG03 A0A251TCA0 A0A251T6N3 A0A0A9WPH5 A0A0K8U6S2 A0A2B4R8Q4 W8AEF0 A0A392M204 A0A0K8U8C3 A0A0V0IV83 A0A151RZS8 A0A085ND65 A0A251UIF2 A0A224XA26 Q6L3N8 A0A0A1XLZ7 W8AQ86 W8AL23 A0A0V0Z6Q0 A0A0A9WRE3 A0A396INR0 A0A0K8U6X2 A0A0A9YB53 A0A0A9W6P7 W8B7K5 A0A0K8WJW9 A0A151T5R0 A0A151TGQ2 A0A2J7PSB2 W8BYI7 A0A1W7R6A5 A0A151THM1 A0A2N9ETM3 A0A0J7K6W1 A0A2N9GX95 A0A2N9IWX1 A0A2N9GK87 Q5MG99 A0A2K3MYT4 A0A151R2G6 A0A2N9HR09 A0A151TGP0 A0A2N9ELX4 A0A2N9IUL9 A0A0V0RWJ3 Q7Y141 A0A151TPU3
Pubmed
EMBL
GEZM01011856
JAV93393.1
GEZM01011855
JAV93396.1
GEZM01011859
JAV93384.1
+ More
GEZM01011857 JAV93390.1 JYDH01000330 KRY26801.1 GDHC01011760 JAQ06869.1 DNYX01000353 HBK84078.1 GBHO01019781 JAG23823.1 KQ460297 KPJ16236.1 GBXI01011094 JAD03198.1 GAKP01015352 JAC43600.1 KQ459875 KPJ19636.1 LBMM01004924 KMQ91985.1 NWSH01000816 PCG74073.1 NEVH01019375 PNF22597.1 NEVH01006980 PNF36330.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 KQ460882 KPJ11482.1 LVZM01021469 OUC41357.1 JYDJ01000808 KRX33413.1 NWSH01002292 PCG68657.1 PCG68658.1 PNF37231.1 JYDJ01000136 KRX42725.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 KL367475 KFD72989.1 KRY26802.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 GBHO01024949 JAG18655.1 GBHO01024951 JAG18653.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 JYDR01000001 KRY80095.1 KL367628 KFD61291.1 JYDU01000001 KRY01876.1 GEZM01045163 GEZM01045162 JAV77888.1 KL367553 KFD64618.1 GEZM01050757 JAV75412.1 CM007901 OTG04806.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 GBXI01017107 JAC97184.1 KA648745 AFP63374.1 CM007899 OTG12557.1 LBMM01008124 KMQ89126.1 CM007900 OTG08534.1 OTG06758.1 GBHO01033237 JAG10367.1 GDHF01030254 JAI22060.1 LSMT01001145 PFX12880.1 GAMC01019995 JAB86560.1 LXQA010002204 MCH81362.1 GDHF01029405 JAI22909.1 GEDG01001927 JAP36462.1 KQ483510 KYP48029.1 KL367515 KFD67411.1 CM007895 OTG22666.1 GFTR01008662 JAW07764.1 AC149291 AAT38758.1 GBXI01002679 JAD11613.1 GAMC01018338 JAB88217.1 GAMC01019998 JAB86557.1 JYDQ01000379 KRY07987.1 GBHO01033235 JAG10369.1 PSQE01000003 RHN67289.1 GDHF01029905 JAI22409.1 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 GAMC01020736 JAB85819.1 GDHF01000873 JAI51441.1 CM003610 KYP62383.1 CM003608 KYP66219.1 NEVH01021933 PNF19227.1 GAMC01008189 JAB98366.1 GEHC01000963 JAV46682.1 KYP66486.1 OIVN01000313 SPC78143.1 LBMM01012645 KMQ86082.1 OIVN01002473 SPD03944.1 OIVN01006239 SPD28653.1 OIVN01002345 SPD02856.1 AY830138 AAV88069.1 ASHM01014005 PNX95963.1 KQ484186 KYP36645.1 OIVN01004297 SPD16537.1 KYP66220.1 OIVN01000172 SPC75650.1 OIVN01006211 SPD27863.1 JYDL01000068 KRX18751.1 AC133339 DP000009 AAP46257.1 ABF99717.1 CM003606 KYP69041.1
GEZM01011857 JAV93390.1 JYDH01000330 KRY26801.1 GDHC01011760 JAQ06869.1 DNYX01000353 HBK84078.1 GBHO01019781 JAG23823.1 KQ460297 KPJ16236.1 GBXI01011094 JAD03198.1 GAKP01015352 JAC43600.1 KQ459875 KPJ19636.1 LBMM01004924 KMQ91985.1 NWSH01000816 PCG74073.1 NEVH01019375 PNF22597.1 NEVH01006980 PNF36330.1 NEVH01002725 PNF41351.1 NEVH01006721 PNF37230.1 KQ460882 KPJ11482.1 LVZM01021469 OUC41357.1 JYDJ01000808 KRX33413.1 NWSH01002292 PCG68657.1 PCG68658.1 PNF37231.1 JYDJ01000136 KRX42725.1 GEZM01013472 GEZM01013471 GEZM01013470 JAV92539.1 KL367475 KFD72989.1 KRY26802.1 JYDT01000069 KRY86576.1 JYDS01000009 JYDV01000021 KRZ33483.1 KRZ41218.1 GBHO01024949 JAG18655.1 GBHO01024951 JAG18653.1 GBHO01024947 JAG18657.1 GBHO01024950 JAG18654.1 JYDR01000001 KRY80095.1 KL367628 KFD61291.1 JYDU01000001 KRY01876.1 GEZM01045163 GEZM01045162 JAV77888.1 KL367553 KFD64618.1 GEZM01050757 JAV75412.1 CM007901 OTG04806.1 GEHC01000867 JAV46778.1 GEHC01000868 JAV46777.1 GBXI01017107 JAC97184.1 KA648745 AFP63374.1 CM007899 OTG12557.1 LBMM01008124 KMQ89126.1 CM007900 OTG08534.1 OTG06758.1 GBHO01033237 JAG10367.1 GDHF01030254 JAI22060.1 LSMT01001145 PFX12880.1 GAMC01019995 JAB86560.1 LXQA010002204 MCH81362.1 GDHF01029405 JAI22909.1 GEDG01001927 JAP36462.1 KQ483510 KYP48029.1 KL367515 KFD67411.1 CM007895 OTG22666.1 GFTR01008662 JAW07764.1 AC149291 AAT38758.1 GBXI01002679 JAD11613.1 GAMC01018338 JAB88217.1 GAMC01019998 JAB86557.1 JYDQ01000379 KRY07987.1 GBHO01033235 JAG10369.1 PSQE01000003 RHN67289.1 GDHF01029905 JAI22409.1 GBHO01016859 JAG26745.1 GBHO01043059 JAG00545.1 GAMC01020736 JAB85819.1 GDHF01000873 JAI51441.1 CM003610 KYP62383.1 CM003608 KYP66219.1 NEVH01021933 PNF19227.1 GAMC01008189 JAB98366.1 GEHC01000963 JAV46682.1 KYP66486.1 OIVN01000313 SPC78143.1 LBMM01012645 KMQ86082.1 OIVN01002473 SPD03944.1 OIVN01006239 SPD28653.1 OIVN01002345 SPD02856.1 AY830138 AAV88069.1 ASHM01014005 PNX95963.1 KQ484186 KYP36645.1 OIVN01004297 SPD16537.1 KYP66220.1 OIVN01000172 SPC75650.1 OIVN01006211 SPD27863.1 JYDL01000068 KRX18751.1 AC133339 DP000009 AAP46257.1 ABF99717.1 CM003606 KYP69041.1
Proteomes
Pfam
Interpro
IPR001878
Znf_CCHC
+ More
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR013103 RVT_2
IPR001584 Integrase_cat-core
IPR039537 Retrotran_Ty1/copia-like
IPR036397 RNaseH_sf
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR011009 Kinase-like_dom_sf
IPR009056 Cyt_c-like_dom
IPR041469 Subtilisin-like_FN3
IPR036852 Peptidase_S8/S53_dom_sf
IPR012337 RNaseH-like_sf
IPR036875 Znf_CCHC_sf
IPR013103 RVT_2
IPR001584 Integrase_cat-core
IPR039537 Retrotran_Ty1/copia-like
IPR036397 RNaseH_sf
IPR025724 GAG-pre-integrase_dom
IPR025314 DUF4219
IPR011009 Kinase-like_dom_sf
IPR009056 Cyt_c-like_dom
IPR041469 Subtilisin-like_FN3
IPR036852 Peptidase_S8/S53_dom_sf
Gene 3D
ProteinModelPortal
A0A1Y1N668
A0A1Y1NAZ7
A0A1Y1N664
A0A1Y1N932
A0A0V1APU2
A0A146LH62
+ More
A0A355BEE2 A0A0A9XY84 A0A194RJV9 A0A0A1WXQ3 A0A034VP70 A0A194RU38 A0A0J7NHJ8 A0A2A4JRI1 A0A2J7Q1Z2 A0A2J7R668 A0A2J7RKJ6 A0A2J7R8S8 A0A0N1ICY1 A0A1Y3EBW8 A0A0V0T3I0 A0A2A4J959 A0A2J7R8R7 A0A0V0TUP0 A0A1Y1N3X2 A0A085NU43 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A0V1F2E6 A0A085MVP5 A0A0V0YNF4 A0A1Y1LYU4 A0A085N572 A0A1Y1LP39 A0A251T2S6 A0A1W7R6K6 A0A1W7R6J6 A0A1X7U495 A0A0A1WFC7 T1PKV2 A0A251TN47 A0A0J7KG03 A0A251TCA0 A0A251T6N3 A0A0A9WPH5 A0A0K8U6S2 A0A2B4R8Q4 W8AEF0 A0A392M204 A0A0K8U8C3 A0A0V0IV83 A0A151RZS8 A0A085ND65 A0A251UIF2 A0A224XA26 Q6L3N8 A0A0A1XLZ7 W8AQ86 W8AL23 A0A0V0Z6Q0 A0A0A9WRE3 A0A396INR0 A0A0K8U6X2 A0A0A9YB53 A0A0A9W6P7 W8B7K5 A0A0K8WJW9 A0A151T5R0 A0A151TGQ2 A0A2J7PSB2 W8BYI7 A0A1W7R6A5 A0A151THM1 A0A2N9ETM3 A0A0J7K6W1 A0A2N9GX95 A0A2N9IWX1 A0A2N9GK87 Q5MG99 A0A2K3MYT4 A0A151R2G6 A0A2N9HR09 A0A151TGP0 A0A2N9ELX4 A0A2N9IUL9 A0A0V0RWJ3 Q7Y141 A0A151TPU3
A0A355BEE2 A0A0A9XY84 A0A194RJV9 A0A0A1WXQ3 A0A034VP70 A0A194RU38 A0A0J7NHJ8 A0A2A4JRI1 A0A2J7Q1Z2 A0A2J7R668 A0A2J7RKJ6 A0A2J7R8S8 A0A0N1ICY1 A0A1Y3EBW8 A0A0V0T3I0 A0A2A4J959 A0A2J7R8R7 A0A0V0TUP0 A0A1Y1N3X2 A0A085NU43 A0A0V1APS7 A0A0V1FKH2 A0A0V1K1Y9 A0A0A9XNJ4 A0A0A9XIP2 A0A0A9XD75 A0A0A9XFC8 A0A0V1F2E6 A0A085MVP5 A0A0V0YNF4 A0A1Y1LYU4 A0A085N572 A0A1Y1LP39 A0A251T2S6 A0A1W7R6K6 A0A1W7R6J6 A0A1X7U495 A0A0A1WFC7 T1PKV2 A0A251TN47 A0A0J7KG03 A0A251TCA0 A0A251T6N3 A0A0A9WPH5 A0A0K8U6S2 A0A2B4R8Q4 W8AEF0 A0A392M204 A0A0K8U8C3 A0A0V0IV83 A0A151RZS8 A0A085ND65 A0A251UIF2 A0A224XA26 Q6L3N8 A0A0A1XLZ7 W8AQ86 W8AL23 A0A0V0Z6Q0 A0A0A9WRE3 A0A396INR0 A0A0K8U6X2 A0A0A9YB53 A0A0A9W6P7 W8B7K5 A0A0K8WJW9 A0A151T5R0 A0A151TGQ2 A0A2J7PSB2 W8BYI7 A0A1W7R6A5 A0A151THM1 A0A2N9ETM3 A0A0J7K6W1 A0A2N9GX95 A0A2N9IWX1 A0A2N9GK87 Q5MG99 A0A2K3MYT4 A0A151R2G6 A0A2N9HR09 A0A151TGP0 A0A2N9ELX4 A0A2N9IUL9 A0A0V0RWJ3 Q7Y141 A0A151TPU3
PDB
1C1A
E-value=0.000389684,
Score=107
Ontologies
KEGG
GO
PANTHER
Topology
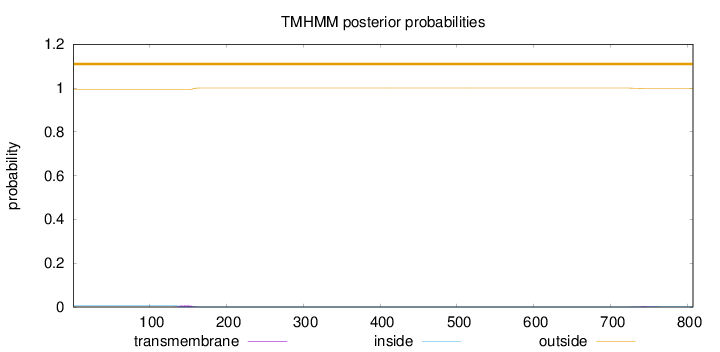
Length:
807
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18756
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00603
outside
1 - 807
Population Genetic Test Statistics
Pi
250.562055
Theta
132.608803
Tajima's D
-0.957149
CLR
0.55644
CSRT
0.144342782860857
Interpretation
Uncertain
