Gene
KWMTBOMO05322 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002521
Annotation
PREDICTED:_gamma-glutamyltransferase_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.809
Sequence
CDS
ATGCAAAGTAAATACGCGCCACAAGGAATGGTGGTGGCACCTCACCACCTTGCTACTCAAAGTGCTATTGATATCCTAAGAGAAGGAGGCACAGCCATAGAGGCAATGGTCGCAGCGGCGGCGACTATATCAGTTGTCTACCCTCATATGAACAGCATTGGAGGTGACTCGTTTTGGCTAATAGTACCACCTCATGGTGATCCCATAGCCATTGAGGCATCCGGCACAGCAGGAAGTTTAGCTACTGAAGATTTCTTCAAAGGCTACGACAAGATTCCGTATCTTGGGCCTAAGGCAGCTATAACCGTTGCGGGCGCTGTCGGAGGTTGGCAGGAAGCCCTAAATTATGTTACCGAAAATGGATACCAGAGGATTTCAGTATCTCGATTATTAGCTGATGCTATAAGGTATGCTGAAGAAGGAGTCTCAGTAACACAAAGTTACATCAGTAATTTAGAAAAATTTAAAGATTTGAATGTATCTAGTGAGTTTCGGAGAATTTATCTATTGGATGGTAACAATCCTAAGATGGGTGATATTTTGTATCAAAAGGAGTTAGGTACTACCTTAAGGCGTCTAGCCGAGAATGGATTAAATAGTTTCTATAAAGGGGAACTAGCAAGAACAATAGCAGAAGACATGGCCACTGTCGGTATGCCGATTACGGAATCTGACTTACAAAACTTTACGGCAATTCGAAAAACTCCCTTGCGACTTTTACTAAAACAAGGAGAAGTTTTCAATCTTCCACCACCTACCCAAGGTTTGGTTTCGTTAAATATCCTTGGAGTGTTAGAAAAACTTAATATAGATGGAAAAAATGAGGGCGAACTTGTGCACGCCACAGTCGAAGCCACAAAGCAAGCTTTTGAAATGAGAAACGATTACATAACTGATCCAAAATATATGAAAATCGATCCCAATTCATTACTGACTAGTCAGCACTTGTCCAAAGTTGCCAGTAAAATAAGTTTGGATAGAGCCGCTTCTGTAGGACAGGGGAAGGGCCCAGGGGACACGATTTGGATGGGGGCTATGGATAGCAAAGGCTTTTCGGTGTCTTTCATTCAAAGTGTATACCACGAGTTTGGAAGTACTGTCGTGTTGCCCAAAACTGGTATACTATGGCAAAATAGAGGAGTTTCTTTCACTTTAGAAAAAGATAAATTATTGACTCTACAGCCTGGAAAAAAACCTTTTCATACGCTAAACTTGGCTACAGCAAGGCTAAATGATGGTAGAGTAATGATATTTGGGACTAGAGGAGGTGACGGACAACCTCAAACCCAATCAGCTATCTTTTACAGACACGTGGTTCAGGGTTTGAACTTACAAGCCGCAGTATCGGCACCGAGATGGCTTTATGGCCGCATCGCAGGGGATGCTGAAGACACCCTGAAAATCGAAAGCAGATTTAGTGAAGAAACCGTGAACTATTTAAAGGAAAGAGGTCACGATGTAGAGATTTTGCCAGAGTTTTCTGAGAAGGTAGGTCACGCTGGTGCATTAGTGAGACACCCGAACGGTGTGATGGAAGGCGCATTCGATCCTCGGTGTGATGGCAGTGCATCGGGATTTTAA
Protein
MQSKYAPQGMVVAPHHLATQSAIDILREGGTAIEAMVAAAATISVVYPHMNSIGGDSFWLIVPPHGDPIAIEASGTAGSLATEDFFKGYDKIPYLGPKAAITVAGAVGGWQEALNYVTENGYQRISVSRLLADAIRYAEEGVSVTQSYISNLEKFKDLNVSSEFRRIYLLDGNNPKMGDILYQKELGTTLRRLAENGLNSFYKGELARTIAEDMATVGMPITESDLQNFTAIRKTPLRLLLKQGEVFNLPPPTQGLVSLNILGVLEKLNIDGKNEGELVHATVEATKQAFEMRNDYITDPKYMKIDPNSLLTSQHLSKVASKISLDRAASVGQGKGPGDTIWMGAMDSKGFSVSFIQSVYHEFGSTVVLPKTGILWQNRGVSFTLEKDKLLTLQPGKKPFHTLNLATARLNDGRVMIFGTRGGDGQPQTQSAIFYRHVVQGLNLQAAVSAPRWLYGRIAGDAEDTLKIESRFSEETVNYLKERGHDVEILPEFSEKVGHAGALVRHPNGVMEGAFDPRCDGSASGF
Summary
Uniprot
H9IZ36
D2Y060
A0A0L7LCL0
A0A2H1VSC5
A0A194QS79
A0A3S2LZ91
+ More
A0A1H4FJW0 A0A3Q0NQT7 A0A1X3RQB4 A0A086G5R0 A0A0N7JP50 A0A221FK59 A0A3G4SN04 A0A2K2IW34 A0A194QKN2 A0A1X3RUS0 A0A383M4R7 A0A3N0UA89 A0A240B4B4 A0A250B8B4 A0A2X2G685 A0A3S4WGH6 A0A1W5DFA7 A0A318NYT0 A0A0X2NT48 A8GA96 A0A2H1LBC2 A0A3M3KFD1 A0A0A3ZKQ3 A0A2I0FSD6 D8MNI1 A0A212FCQ0 A0A379YZW9 A0A2I5THH1 A0A3R2C815 S4YF62 A0A3R2EIR2 A0A3S4E4L4 D4DWQ4 A0A0X8SH01 A0A380A0M9 A0A1X0WCN1 A0A1B1KLJ6 A0A2T4XZX2 A0A1V2QZW6 A0A389MDK1 A0A0Q4LXN5 A0A1X1ERE5 A0A286BRA0 A0A1X0W682 J3HL28 A0A0U5KYK8 A0A3M7UK50 A0A3D0ERD2 A0A3Q8HAT0 A0A2V1YIR4 A0A2V5BX72 A0A2S9IH10 J2LGC3 A0A1V9DBN9 A0A366N5S2 A0A0L7T2H7 I0QPM2 E1SEH5 U3TN54 A0A3A3ZX21 A0A2U9DCN6 A0A2S8A0K5 A0A1T4JIB3 A0A330JA64 A0A093US13 A0A0H3F5R8 H8NPN3 U2MY69 A0A0Q4MBR8 A0A2N0MY80 A0A2R4J595 A0A2W4CPN3 A0A3N1W050 A0A221T672 A0A0F5Y677 A0A377NGX6 E0LUQ7 A0A085GMP0 A0A0J8YHF9 A0A1X1DK82 A0A0B2SZX9 A0A2G8DB52 A0A1M5TXC8 A0A0M2EVL0 A0A1X1DAG6 A0A1I5AVK2 A0A1D7Z2W5 A0A094RS46 A0A0T9M539 A0A0A3YYS6 A0A2V3SLW0 A0A3R8WY56 A0A3N6UL45 A0A086ES50
A0A1H4FJW0 A0A3Q0NQT7 A0A1X3RQB4 A0A086G5R0 A0A0N7JP50 A0A221FK59 A0A3G4SN04 A0A2K2IW34 A0A194QKN2 A0A1X3RUS0 A0A383M4R7 A0A3N0UA89 A0A240B4B4 A0A250B8B4 A0A2X2G685 A0A3S4WGH6 A0A1W5DFA7 A0A318NYT0 A0A0X2NT48 A8GA96 A0A2H1LBC2 A0A3M3KFD1 A0A0A3ZKQ3 A0A2I0FSD6 D8MNI1 A0A212FCQ0 A0A379YZW9 A0A2I5THH1 A0A3R2C815 S4YF62 A0A3R2EIR2 A0A3S4E4L4 D4DWQ4 A0A0X8SH01 A0A380A0M9 A0A1X0WCN1 A0A1B1KLJ6 A0A2T4XZX2 A0A1V2QZW6 A0A389MDK1 A0A0Q4LXN5 A0A1X1ERE5 A0A286BRA0 A0A1X0W682 J3HL28 A0A0U5KYK8 A0A3M7UK50 A0A3D0ERD2 A0A3Q8HAT0 A0A2V1YIR4 A0A2V5BX72 A0A2S9IH10 J2LGC3 A0A1V9DBN9 A0A366N5S2 A0A0L7T2H7 I0QPM2 E1SEH5 U3TN54 A0A3A3ZX21 A0A2U9DCN6 A0A2S8A0K5 A0A1T4JIB3 A0A330JA64 A0A093US13 A0A0H3F5R8 H8NPN3 U2MY69 A0A0Q4MBR8 A0A2N0MY80 A0A2R4J595 A0A2W4CPN3 A0A3N1W050 A0A221T672 A0A0F5Y677 A0A377NGX6 E0LUQ7 A0A085GMP0 A0A0J8YHF9 A0A1X1DK82 A0A0B2SZX9 A0A2G8DB52 A0A1M5TXC8 A0A0M2EVL0 A0A1X1DAG6 A0A1I5AVK2 A0A1D7Z2W5 A0A094RS46 A0A0T9M539 A0A0A3YYS6 A0A2V3SLW0 A0A3R8WY56 A0A3N6UL45 A0A086ES50
Pubmed
EMBL
BABH01011016
GU270851
ADB27113.1
JTDY01001696
KOB73135.1
ODYU01004154
+ More
SOQ43719.1 KQ461154 KPJ08378.1 RSAL01000103 RVE47434.1 FNQS01000013 SEA97337.1 LCWI01000010 KKZ18146.1 LUTO01000052 OSN03972.1 JPUX01000002 KFF86524.1 CP013046 ALL36638.1 CP018926 ASM10590.1 CP033504 AYU89020.1 PPGF01000005 PNU39760.1 KQ458714 KPJ05475.1 LUTP01000020 OSN05620.1 UIRQ01000170 SVK46869.1 RJUJ01000015 ROH77480.1 LT906479 SNV89868.1 CP014136 ATA22384.1 UAUM01000002 SPZ52892.1 LR134494 VEI65150.1 FWWG01000004 SMB24613.1 PESE01000005 PYD38028.1 FBSD01000008 CUW03428.1 CP000826 ABV40036.1 LT883155 SMZ55240.1 RBOX01000029 RMN21758.1 JRUQ01000102 KGT86333.1 NVXY01000012 PKH21230.1 FP236843 CAX58388.1 AGBW02009158 OWR51493.1 UGYN01000002 SUI52927.1 CP025084 AUH04011.1 RCUM01000284 MNG64574.1 CP006566 AGP43140.1 RCUE01000003 RCUO01000007 RCUM01000213 RCUN01000006 RCUS01000023 MMZ40001.1 MNB56459.1 MNG62108.1 MNH52291.1 MNL95564.1 LR134117 VDZ52187.1 ADBY01000013 EFE98026.1 CP014017 AMG99132.1 UGYL01000001 SUI72010.1 MRWE01000027 ORJ24547.1 CP015613 ANS41318.1 PZPP01000013 PTM35489.1 MPUJ01000015 ONK02466.1 BHES01000073 GBU13570.1 LMLI01000019 KQN44125.1 MLJI01000001 ORM92579.1 OCMY01000001 SOD36671.1 MRWD01000009 ORJ22296.1 AKIT01000038 EJM98557.1 LN907827 CUU23086.1 RHDS01000002 RNA78187.1 DOTQ01000045 HBZ17694.1 CP022725 AXU96262.1 QGHE01000016 PWJ74190.1 QKMJ01000008 PYG47686.1 PDET01000001 PRD17073.1 AKIU01000026 EJL91265.1 MWUE01000028 OQP31195.1 JRXE01000015 KOC89634.1 AJHJ01000027 EIC83245.1 CP002206 ADO08466.1 AP012551 BAN95140.1 QZDF01000039 RJL19148.1 CP020820 AWP33893.1 PUEK01000002 PQL29028.1 FUWI01000001 SJZ29912.1 OANP01000051 SNW60808.1 JQHN01000022 KFX11008.1 CP002505 ADW72556.1 CP003403 AFE57137.1 ASZC01000026 ERK06540.1 LMLJ01000055 KQN49037.1 LXYV01000788 PKB87265.1 CP028349 AVV36802.1 PUFZ01000033 PZL94865.1 RJVB01000007 ROR06748.1 CP021894 ASN84399.1 JMRT02000001 KKD34278.1 UGGO01000001 STQ46038.1 AEDL01000002 EFM21033.1 JMPJ01000022 KFC84985.1 JMSO01000178 KMV69227.1 MIPO01000145 ORM76960.1 JSXC01000039 KHN50498.1 MTCH01000013 PIJ59527.1 FQWI01000012 SHH55379.1 JQOD01000013 KGA31269.1 MLFS01000018 ORM73634.1 FOVG01000001 SFN66455.1 CP015750 AOR64198.1 JQOF01000043 KGA39255.1 CPYI01000025 CNF59764.1 JRUP01000015 KGT90486.1 QICZ01000001 PXW21282.1 RHHM01000018 RQM36659.1 JPSO01000047 KFF69514.1
SOQ43719.1 KQ461154 KPJ08378.1 RSAL01000103 RVE47434.1 FNQS01000013 SEA97337.1 LCWI01000010 KKZ18146.1 LUTO01000052 OSN03972.1 JPUX01000002 KFF86524.1 CP013046 ALL36638.1 CP018926 ASM10590.1 CP033504 AYU89020.1 PPGF01000005 PNU39760.1 KQ458714 KPJ05475.1 LUTP01000020 OSN05620.1 UIRQ01000170 SVK46869.1 RJUJ01000015 ROH77480.1 LT906479 SNV89868.1 CP014136 ATA22384.1 UAUM01000002 SPZ52892.1 LR134494 VEI65150.1 FWWG01000004 SMB24613.1 PESE01000005 PYD38028.1 FBSD01000008 CUW03428.1 CP000826 ABV40036.1 LT883155 SMZ55240.1 RBOX01000029 RMN21758.1 JRUQ01000102 KGT86333.1 NVXY01000012 PKH21230.1 FP236843 CAX58388.1 AGBW02009158 OWR51493.1 UGYN01000002 SUI52927.1 CP025084 AUH04011.1 RCUM01000284 MNG64574.1 CP006566 AGP43140.1 RCUE01000003 RCUO01000007 RCUM01000213 RCUN01000006 RCUS01000023 MMZ40001.1 MNB56459.1 MNG62108.1 MNH52291.1 MNL95564.1 LR134117 VDZ52187.1 ADBY01000013 EFE98026.1 CP014017 AMG99132.1 UGYL01000001 SUI72010.1 MRWE01000027 ORJ24547.1 CP015613 ANS41318.1 PZPP01000013 PTM35489.1 MPUJ01000015 ONK02466.1 BHES01000073 GBU13570.1 LMLI01000019 KQN44125.1 MLJI01000001 ORM92579.1 OCMY01000001 SOD36671.1 MRWD01000009 ORJ22296.1 AKIT01000038 EJM98557.1 LN907827 CUU23086.1 RHDS01000002 RNA78187.1 DOTQ01000045 HBZ17694.1 CP022725 AXU96262.1 QGHE01000016 PWJ74190.1 QKMJ01000008 PYG47686.1 PDET01000001 PRD17073.1 AKIU01000026 EJL91265.1 MWUE01000028 OQP31195.1 JRXE01000015 KOC89634.1 AJHJ01000027 EIC83245.1 CP002206 ADO08466.1 AP012551 BAN95140.1 QZDF01000039 RJL19148.1 CP020820 AWP33893.1 PUEK01000002 PQL29028.1 FUWI01000001 SJZ29912.1 OANP01000051 SNW60808.1 JQHN01000022 KFX11008.1 CP002505 ADW72556.1 CP003403 AFE57137.1 ASZC01000026 ERK06540.1 LMLJ01000055 KQN49037.1 LXYV01000788 PKB87265.1 CP028349 AVV36802.1 PUFZ01000033 PZL94865.1 RJVB01000007 ROR06748.1 CP021894 ASN84399.1 JMRT02000001 KKD34278.1 UGGO01000001 STQ46038.1 AEDL01000002 EFM21033.1 JMPJ01000022 KFC84985.1 JMSO01000178 KMV69227.1 MIPO01000145 ORM76960.1 JSXC01000039 KHN50498.1 MTCH01000013 PIJ59527.1 FQWI01000012 SHH55379.1 JQOD01000013 KGA31269.1 MLFS01000018 ORM73634.1 FOVG01000001 SFN66455.1 CP015750 AOR64198.1 JQOF01000043 KGA39255.1 CPYI01000025 CNF59764.1 JRUP01000015 KGT90486.1 QICZ01000001 PXW21282.1 RHHM01000018 RQM36659.1 JPSO01000047 KFF69514.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000283053
UP000187280
UP000034361
+ More
UP000194062 UP000028708 UP000058492 UP000198289 UP000281251 UP000236556 UP000053268 UP000194020 UP000258172 UP000274511 UP000215134 UP000217182 UP000250791 UP000273164 UP000192912 UP000248196 UP000242817 UP000007074 UP000246092 UP000030351 UP000237543 UP000008793 UP000007151 UP000255529 UP000017700 UP000014900 UP000281391 UP000005723 UP000062490 UP000254742 UP000192536 UP000013567 UP000241614 UP000189286 UP000282082 UP000051594 UP000193749 UP000219271 UP000192722 UP000007506 UP000059419 UP000278924 UP000266781 UP000245996 UP000247871 UP000239181 UP000007295 UP000192769 UP000037088 UP000011072 UP000006631 UP000016901 UP000266354 UP000247725 UP000238919 UP000242630 UP000250713 UP000033091 UP000007257 UP000007594 UP000016607 UP000050990 UP000232559 UP000241538 UP000249374 UP000275666 UP000263924 UP000033668 UP000254304 UP000004279 UP000028640 UP000037152 UP000193803 UP000053038 UP000229180 UP000184537 UP000029435 UP000193104 UP000198968 UP000094471 UP000029447 UP000045824 UP000030397 UP000248397 UP000279457 UP000032862
UP000194062 UP000028708 UP000058492 UP000198289 UP000281251 UP000236556 UP000053268 UP000194020 UP000258172 UP000274511 UP000215134 UP000217182 UP000250791 UP000273164 UP000192912 UP000248196 UP000242817 UP000007074 UP000246092 UP000030351 UP000237543 UP000008793 UP000007151 UP000255529 UP000017700 UP000014900 UP000281391 UP000005723 UP000062490 UP000254742 UP000192536 UP000013567 UP000241614 UP000189286 UP000282082 UP000051594 UP000193749 UP000219271 UP000192722 UP000007506 UP000059419 UP000278924 UP000266781 UP000245996 UP000247871 UP000239181 UP000007295 UP000192769 UP000037088 UP000011072 UP000006631 UP000016901 UP000266354 UP000247725 UP000238919 UP000242630 UP000250713 UP000033091 UP000007257 UP000007594 UP000016607 UP000050990 UP000232559 UP000241538 UP000249374 UP000275666 UP000263924 UP000033668 UP000254304 UP000004279 UP000028640 UP000037152 UP000193803 UP000053038 UP000229180 UP000184537 UP000029435 UP000193104 UP000198968 UP000094471 UP000029447 UP000045824 UP000030397 UP000248397 UP000279457 UP000032862
Interpro
IPR029055
Ntn_hydrolases_N
SUPFAM
SSF56235
SSF56235
ProteinModelPortal
H9IZ36
D2Y060
A0A0L7LCL0
A0A2H1VSC5
A0A194QS79
A0A3S2LZ91
+ More
A0A1H4FJW0 A0A3Q0NQT7 A0A1X3RQB4 A0A086G5R0 A0A0N7JP50 A0A221FK59 A0A3G4SN04 A0A2K2IW34 A0A194QKN2 A0A1X3RUS0 A0A383M4R7 A0A3N0UA89 A0A240B4B4 A0A250B8B4 A0A2X2G685 A0A3S4WGH6 A0A1W5DFA7 A0A318NYT0 A0A0X2NT48 A8GA96 A0A2H1LBC2 A0A3M3KFD1 A0A0A3ZKQ3 A0A2I0FSD6 D8MNI1 A0A212FCQ0 A0A379YZW9 A0A2I5THH1 A0A3R2C815 S4YF62 A0A3R2EIR2 A0A3S4E4L4 D4DWQ4 A0A0X8SH01 A0A380A0M9 A0A1X0WCN1 A0A1B1KLJ6 A0A2T4XZX2 A0A1V2QZW6 A0A389MDK1 A0A0Q4LXN5 A0A1X1ERE5 A0A286BRA0 A0A1X0W682 J3HL28 A0A0U5KYK8 A0A3M7UK50 A0A3D0ERD2 A0A3Q8HAT0 A0A2V1YIR4 A0A2V5BX72 A0A2S9IH10 J2LGC3 A0A1V9DBN9 A0A366N5S2 A0A0L7T2H7 I0QPM2 E1SEH5 U3TN54 A0A3A3ZX21 A0A2U9DCN6 A0A2S8A0K5 A0A1T4JIB3 A0A330JA64 A0A093US13 A0A0H3F5R8 H8NPN3 U2MY69 A0A0Q4MBR8 A0A2N0MY80 A0A2R4J595 A0A2W4CPN3 A0A3N1W050 A0A221T672 A0A0F5Y677 A0A377NGX6 E0LUQ7 A0A085GMP0 A0A0J8YHF9 A0A1X1DK82 A0A0B2SZX9 A0A2G8DB52 A0A1M5TXC8 A0A0M2EVL0 A0A1X1DAG6 A0A1I5AVK2 A0A1D7Z2W5 A0A094RS46 A0A0T9M539 A0A0A3YYS6 A0A2V3SLW0 A0A3R8WY56 A0A3N6UL45 A0A086ES50
A0A1H4FJW0 A0A3Q0NQT7 A0A1X3RQB4 A0A086G5R0 A0A0N7JP50 A0A221FK59 A0A3G4SN04 A0A2K2IW34 A0A194QKN2 A0A1X3RUS0 A0A383M4R7 A0A3N0UA89 A0A240B4B4 A0A250B8B4 A0A2X2G685 A0A3S4WGH6 A0A1W5DFA7 A0A318NYT0 A0A0X2NT48 A8GA96 A0A2H1LBC2 A0A3M3KFD1 A0A0A3ZKQ3 A0A2I0FSD6 D8MNI1 A0A212FCQ0 A0A379YZW9 A0A2I5THH1 A0A3R2C815 S4YF62 A0A3R2EIR2 A0A3S4E4L4 D4DWQ4 A0A0X8SH01 A0A380A0M9 A0A1X0WCN1 A0A1B1KLJ6 A0A2T4XZX2 A0A1V2QZW6 A0A389MDK1 A0A0Q4LXN5 A0A1X1ERE5 A0A286BRA0 A0A1X0W682 J3HL28 A0A0U5KYK8 A0A3M7UK50 A0A3D0ERD2 A0A3Q8HAT0 A0A2V1YIR4 A0A2V5BX72 A0A2S9IH10 J2LGC3 A0A1V9DBN9 A0A366N5S2 A0A0L7T2H7 I0QPM2 E1SEH5 U3TN54 A0A3A3ZX21 A0A2U9DCN6 A0A2S8A0K5 A0A1T4JIB3 A0A330JA64 A0A093US13 A0A0H3F5R8 H8NPN3 U2MY69 A0A0Q4MBR8 A0A2N0MY80 A0A2R4J595 A0A2W4CPN3 A0A3N1W050 A0A221T672 A0A0F5Y677 A0A377NGX6 E0LUQ7 A0A085GMP0 A0A0J8YHF9 A0A1X1DK82 A0A0B2SZX9 A0A2G8DB52 A0A1M5TXC8 A0A0M2EVL0 A0A1X1DAG6 A0A1I5AVK2 A0A1D7Z2W5 A0A094RS46 A0A0T9M539 A0A0A3YYS6 A0A2V3SLW0 A0A3R8WY56 A0A3N6UL45 A0A086ES50
PDB
5HFT
E-value=9.06878e-82,
Score=774
Ontologies
GO
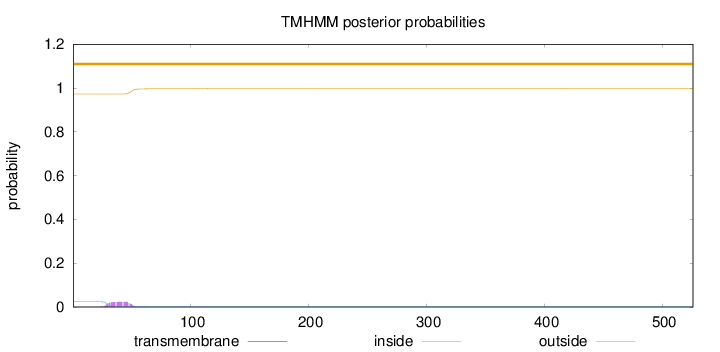
Topology
Length:
526
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.525449999999999
Exp number, first 60 AAs:
0.49912
Total prob of N-in:
0.02665
outside
1 - 526
Population Genetic Test Statistics
Pi
273.317873
Theta
195.489288
Tajima's D
1.212182
CLR
0.000362
CSRT
0.71886405679716
Interpretation
Uncertain
