Gene
KWMTBOMO05318 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002394
Annotation
PREDICTED:_methionine_aminopeptidase_1_isoform_X1_[Bombyx_mori]
Full name
Methionine aminopeptidase
Location in the cell
Cytoplasmic Reliability : 2.032
Sequence
CDS
ATGGCGACGCTTGGAATGTGCGAAACTCCCGGGTGCAAATCAATCGCACAATTACAATGCCCTACTTGTATCAAACTCGGGATTAAAGGCTCGTTCTTTTGTAACCAAGACTGTTTCAAAAAATCGTGGAAGACTCATAAAATAATTCATTCATTAGCGAAAGGTGACAATACCGATGCTGCAAACGTGGAATATAACCCATGGCCGTCGTATACCTTTACGGGAAAGCTGAGACCGTTCCCGGCAGGGCCAAAACGCACTGTGCCGCCTCATATAGGTCGTCCGGATTACGCTGATCATCCGACCGGGTTCCCGGCTTCAGAGTATGCTGCTAAAGGTTCAGCTCAAATCAAGATCCTTGACGATGAAGAGATTGAGGGCATGCGTGTTGCTTGCCGCCTCGGCAGAGAAGTTCTGGATGAAGCAGCTAGAGTGTGTGATGTAGGAGTTAGCACAGATGAGATTGACCGTGTAGTCCACGAGGCTTGCATTGAACGGGAATGTTACCCAAGTCCCCTGAATTATTATAATTTCCCCAACAGTTGTTGTACTTCAATTAATGAGGTTATATGTCATGGAATACCAGATTTACGTCCATTACAGGATGGGGACATATGCAATGTTGATGTAACAGTTTACCATAGAGGTTTCCATGGTGATCTAAATGAAACCTTTTTTGTTGGGAATGTACCAGAGACTACTCGTAAATTAGTTCAGGTCACCCATGAATGCTTACAGAAAGCCATAGATATAGTCCGTCCTGGTGAAAAGTACAGGGAGATTGGCAATGTAATACAGAAACATGCTCAAGCAAATGGATATAGTGTAGTAAGATCTTATTGTGGTCATGGCATACATAGATTGTTTCATACAGCTCCAAATGTACCACATTATGCTAAAAACAAAGCTATTGGTATTATGAAAGTAGGACATTGTTTCACAATAGAACCAATGATCAATGAAGGAGGCTGGCGGGACGAGCAGTGGCCCGATCACTGGACAGCCGTTACCGCTGATGGATCAAGATCTGCACAGTTTGAACAAACCCTTCTAGTAACAGAATCGGGCTGTGACATTTTAACCAAACGTAACACGGGAAGACCCTGGTTCCTTGATCAACTCGAGAAAATTGAAGCGAAGGATAAGTCTTAG
Protein
MATLGMCETPGCKSIAQLQCPTCIKLGIKGSFFCNQDCFKKSWKTHKIIHSLAKGDNTDAANVEYNPWPSYTFTGKLRPFPAGPKRTVPPHIGRPDYADHPTGFPASEYAAKGSAQIKILDDEEIEGMRVACRLGREVLDEAARVCDVGVSTDEIDRVVHEACIERECYPSPLNYYNFPNSCCTSINEVICHGIPDLRPLQDGDICNVDVTVYHRGFHGDLNETFFVGNVPETTRKLVQVTHECLQKAIDIVRPGEKYREIGNVIQKHAQANGYSVVRSYCGHGIHRLFHTAPNVPHYAKNKAIGIMKVGHCFTIEPMINEGGWRDEQWPDHWTAVTADGSRSAQFEQTLLVTESGCDILTKRNTGRPWFLDQLEKIEAKDKS
Summary
Description
Removes the N-terminal methionine from nascent proteins. The N-terminal methionine is often cleaved when the second residue in the primary sequence is small and uncharged (Met-Ala-, Cys, Gly, Pro, Ser, Thr, or Val).
Catalytic Activity
Release of N-terminal amino acids, preferentially methionine, from peptides and arylamides.
Similarity
Belongs to the peptidase M24A family. Methionine aminopeptidase type 1 subfamily.
Belongs to the peptidase M24A family.
Belongs to the peptidase M24A family.
Uniprot
H9IYR0
A0A2A4K1K6
S4P7N6
A0A2H1VWB4
A0A194QBM5
A0A212EJW8
+ More
A0A088AK91 D6X4P7 A0A0L7QQE3 A0A0M8ZVR5 A0A2A3ECH8 A0A1Y1NMM5 A0A0C9RES9 A0A026W4M4 A0A0T6B8S3 F4WU86 A0A158NTW4 A0A195C854 A0A232F417 A0A151JQQ9 K7IR17 E9IH60 E2BLG2 A0A182PTT1 A0A182W2Z1 A0A310SLT2 A0A182JYB7 A0A182WVF8 A0A182R8C9 A0A182I937 A0A182M4K1 R4WK40 A0A182YEU7 A0A182QXY3 A0A154PEU5 A0A182NCK4 A0A2J7RI72 A0A182KXY0 Q7QAH2 A0A182GP12 A0A182VLA5 Q0IFY6 A0A084WR40 E0VK43 A0A182JMZ4 A0A182SEW5 A0A1B6GC21 A0A1B6HYE0 B0WEW3 A0A182TJU8 A0A069DTH1 A0A182F8E1 A0A0A9Y188 A0A0P4VR59 A0A2M4BRK8 R4G8A5 W5JJK1 A0A1Q3FE60 A0A1B6LG69 A0A1B6CLC8 A0A2M4AQQ9 A0A0K8SLU0 A0A224YNV7 A0A131YSJ1 A0A194QSB9 A0A131X9M3 A0A023GJG4 G3MNE0 A0A023FKV8 A0A1B0C8Y0 U4USJ6 A0A1L8E2P3 L7M8Z5 A0A0P5K4P4 A0A1Z5L5F0 A0A0P6ASA1 A0A2R5LM79 A0A0P5G1E8 A0A0P5T2W0 A0A1S3D0B2 A0A336L8V3 A0A1E1X561 A0A0K8RD90 A0A293MTK8 U5EZB0 A0A0C9SCA6 A0A1W4W2P6 E2AWT8 E9GAH7 A0A1A9WC32 B4PV57 A0A067QZU9 B4HH59 T1G1H4 Q9VC48 A0A210R4V1 B3M040 A0A1B0A8U0 T1IS65 A0A034WMP3
A0A088AK91 D6X4P7 A0A0L7QQE3 A0A0M8ZVR5 A0A2A3ECH8 A0A1Y1NMM5 A0A0C9RES9 A0A026W4M4 A0A0T6B8S3 F4WU86 A0A158NTW4 A0A195C854 A0A232F417 A0A151JQQ9 K7IR17 E9IH60 E2BLG2 A0A182PTT1 A0A182W2Z1 A0A310SLT2 A0A182JYB7 A0A182WVF8 A0A182R8C9 A0A182I937 A0A182M4K1 R4WK40 A0A182YEU7 A0A182QXY3 A0A154PEU5 A0A182NCK4 A0A2J7RI72 A0A182KXY0 Q7QAH2 A0A182GP12 A0A182VLA5 Q0IFY6 A0A084WR40 E0VK43 A0A182JMZ4 A0A182SEW5 A0A1B6GC21 A0A1B6HYE0 B0WEW3 A0A182TJU8 A0A069DTH1 A0A182F8E1 A0A0A9Y188 A0A0P4VR59 A0A2M4BRK8 R4G8A5 W5JJK1 A0A1Q3FE60 A0A1B6LG69 A0A1B6CLC8 A0A2M4AQQ9 A0A0K8SLU0 A0A224YNV7 A0A131YSJ1 A0A194QSB9 A0A131X9M3 A0A023GJG4 G3MNE0 A0A023FKV8 A0A1B0C8Y0 U4USJ6 A0A1L8E2P3 L7M8Z5 A0A0P5K4P4 A0A1Z5L5F0 A0A0P6ASA1 A0A2R5LM79 A0A0P5G1E8 A0A0P5T2W0 A0A1S3D0B2 A0A336L8V3 A0A1E1X561 A0A0K8RD90 A0A293MTK8 U5EZB0 A0A0C9SCA6 A0A1W4W2P6 E2AWT8 E9GAH7 A0A1A9WC32 B4PV57 A0A067QZU9 B4HH59 T1G1H4 Q9VC48 A0A210R4V1 B3M040 A0A1B0A8U0 T1IS65 A0A034WMP3
EC Number
3.4.11.18
Pubmed
19121390
23622113
26354079
22118469
18362917
19820115
+ More
28004739 24508170 30249741 21719571 21347285 28648823 20075255 21282665 20798317 23691247 25244985 20966253 12364791 14747013 17210077 26483478 17510324 24438588 20566863 26334808 25401762 26823975 27129103 20920257 23761445 28797301 26830274 28049606 22216098 23537049 25576852 28528879 28503490 26131772 21292972 17994087 17550304 24845553 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 25348373
28004739 24508170 30249741 21719571 21347285 28648823 20075255 21282665 20798317 23691247 25244985 20966253 12364791 14747013 17210077 26483478 17510324 24438588 20566863 26334808 25401762 26823975 27129103 20920257 23761445 28797301 26830274 28049606 22216098 23537049 25576852 28528879 28503490 26131772 21292972 17994087 17550304 24845553 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28812685 25348373
EMBL
BABH01010999
NWSH01000234
PCG78151.1
GAIX01004509
JAA88051.1
ODYU01004846
+ More
SOQ45141.1 KQ459220 KPJ02948.1 AGBW02014362 OWR41787.1 KQ971380 EEZ97675.1 KQ414792 KOC60741.1 KQ435827 KOX71937.1 KZ288285 PBC29445.1 GEZM01002596 JAV97197.1 GBYB01015104 JAG84871.1 KK107423 QOIP01000009 EZA51002.1 RLU18315.1 LJIG01009113 KRT83730.1 GL888355 EGI62194.1 ADTU01026172 KQ978143 KYM96830.1 NNAY01001082 OXU25210.1 KQ978646 KYN29450.1 GL763174 EFZ20105.1 GL449017 EFN83514.1 KQ761453 OAD57539.1 AXCM01000016 AK417975 BAN21190.1 AXCN02001184 KQ434889 KZC10361.1 NEVH01003505 PNF40531.1 AAAB01008888 EAA08766.5 JXUM01077428 KQ563008 KXJ74647.1 CH477281 EAT44938.1 ATLV01025918 KE525402 KFB52684.1 DS235239 EEB13749.1 GECZ01009782 JAS59987.1 GECU01028022 JAS79684.1 DS231912 EDS25821.1 GBGD01001828 JAC87061.1 GBHO01020334 GDHC01017032 JAG23270.1 JAQ01597.1 GDKW01001567 JAI55028.1 GGFJ01006472 MBW55613.1 ACPB03018778 GAHY01001312 JAA76198.1 ADMH02001252 ETN63473.1 GFDL01009200 JAV25845.1 GEBQ01017413 JAT22564.1 GEDC01023110 JAS14188.1 GGFK01009806 MBW43127.1 GBRD01011512 JAG54312.1 GFPF01004396 MAA15542.1 GEDV01006333 JAP82224.1 KQ461154 KPJ08382.1 GEFH01004287 JAP64294.1 GBBM01001359 JAC34059.1 JO843391 AEO35008.1 GBBK01002688 JAC21794.1 AJWK01001597 KB632345 ERL93075.1 GFDF01001097 JAV12987.1 GACK01004687 JAA60347.1 GDIQ01190970 LRGB01002019 JAK60755.1 KZS09631.1 GFJQ02004470 JAW02500.1 GDIP01025116 JAM78599.1 GGLE01006488 MBY10614.1 GDIQ01250912 JAK00813.1 GDIP01131800 JAL71914.1 UFQS01002356 UFQT01002356 SSX13859.1 SSX33279.1 GFAC01004786 JAT94402.1 GADI01004701 JAA69107.1 GFWV01018711 MAA43439.1 GANO01001119 JAB58752.1 GBZX01001433 JAG91307.1 GL443425 EFN62058.1 GL732537 EFX83524.1 CM000160 EDW98906.1 KK852809 KDR15975.1 CH480815 EDW43535.1 AMQM01002988 KB095959 ESO09558.1 AE014297 AY058368 AAF56327.1 AAL13597.1 NEDP02000449 OWF55934.1 CH902617 EDV41997.2 JH431424 GAKP01002136 GAKP01002135 JAC56816.1
SOQ45141.1 KQ459220 KPJ02948.1 AGBW02014362 OWR41787.1 KQ971380 EEZ97675.1 KQ414792 KOC60741.1 KQ435827 KOX71937.1 KZ288285 PBC29445.1 GEZM01002596 JAV97197.1 GBYB01015104 JAG84871.1 KK107423 QOIP01000009 EZA51002.1 RLU18315.1 LJIG01009113 KRT83730.1 GL888355 EGI62194.1 ADTU01026172 KQ978143 KYM96830.1 NNAY01001082 OXU25210.1 KQ978646 KYN29450.1 GL763174 EFZ20105.1 GL449017 EFN83514.1 KQ761453 OAD57539.1 AXCM01000016 AK417975 BAN21190.1 AXCN02001184 KQ434889 KZC10361.1 NEVH01003505 PNF40531.1 AAAB01008888 EAA08766.5 JXUM01077428 KQ563008 KXJ74647.1 CH477281 EAT44938.1 ATLV01025918 KE525402 KFB52684.1 DS235239 EEB13749.1 GECZ01009782 JAS59987.1 GECU01028022 JAS79684.1 DS231912 EDS25821.1 GBGD01001828 JAC87061.1 GBHO01020334 GDHC01017032 JAG23270.1 JAQ01597.1 GDKW01001567 JAI55028.1 GGFJ01006472 MBW55613.1 ACPB03018778 GAHY01001312 JAA76198.1 ADMH02001252 ETN63473.1 GFDL01009200 JAV25845.1 GEBQ01017413 JAT22564.1 GEDC01023110 JAS14188.1 GGFK01009806 MBW43127.1 GBRD01011512 JAG54312.1 GFPF01004396 MAA15542.1 GEDV01006333 JAP82224.1 KQ461154 KPJ08382.1 GEFH01004287 JAP64294.1 GBBM01001359 JAC34059.1 JO843391 AEO35008.1 GBBK01002688 JAC21794.1 AJWK01001597 KB632345 ERL93075.1 GFDF01001097 JAV12987.1 GACK01004687 JAA60347.1 GDIQ01190970 LRGB01002019 JAK60755.1 KZS09631.1 GFJQ02004470 JAW02500.1 GDIP01025116 JAM78599.1 GGLE01006488 MBY10614.1 GDIQ01250912 JAK00813.1 GDIP01131800 JAL71914.1 UFQS01002356 UFQT01002356 SSX13859.1 SSX33279.1 GFAC01004786 JAT94402.1 GADI01004701 JAA69107.1 GFWV01018711 MAA43439.1 GANO01001119 JAB58752.1 GBZX01001433 JAG91307.1 GL443425 EFN62058.1 GL732537 EFX83524.1 CM000160 EDW98906.1 KK852809 KDR15975.1 CH480815 EDW43535.1 AMQM01002988 KB095959 ESO09558.1 AE014297 AY058368 AAF56327.1 AAL13597.1 NEDP02000449 OWF55934.1 CH902617 EDV41997.2 JH431424 GAKP01002136 GAKP01002135 JAC56816.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000005203
UP000007266
+ More
UP000053825 UP000053105 UP000242457 UP000053097 UP000279307 UP000007755 UP000005205 UP000078542 UP000215335 UP000078492 UP000002358 UP000008237 UP000075885 UP000075920 UP000075881 UP000075900 UP000075883 UP000076408 UP000075886 UP000076502 UP000075884 UP000235965 UP000075882 UP000007062 UP000069940 UP000249989 UP000075903 UP000008820 UP000030765 UP000009046 UP000075880 UP000075901 UP000002320 UP000075902 UP000069272 UP000015103 UP000000673 UP000053240 UP000092461 UP000030742 UP000076858 UP000079169 UP000192221 UP000000311 UP000000305 UP000091820 UP000002282 UP000027135 UP000001292 UP000015101 UP000000803 UP000242188 UP000007801 UP000092445
UP000053825 UP000053105 UP000242457 UP000053097 UP000279307 UP000007755 UP000005205 UP000078542 UP000215335 UP000078492 UP000002358 UP000008237 UP000075885 UP000075920 UP000075881 UP000075900 UP000075883 UP000076408 UP000075886 UP000076502 UP000075884 UP000235965 UP000075882 UP000007062 UP000069940 UP000249989 UP000075903 UP000008820 UP000030765 UP000009046 UP000075880 UP000075901 UP000002320 UP000075902 UP000069272 UP000015103 UP000000673 UP000053240 UP000092461 UP000030742 UP000076858 UP000079169 UP000192221 UP000000311 UP000000305 UP000091820 UP000002282 UP000027135 UP000001292 UP000015101 UP000000803 UP000242188 UP000007801 UP000092445
Pfam
Interpro
IPR031615
Zfn-C6H2
+ More
IPR001714 Pept_M24_MAP
IPR002467 Pept_M24A_MAP1
IPR000994 Pept_M24
IPR036005 Creatinase/aminopeptidase-like
IPR007235 Glyco_trans_28_C
IPR007034 BMS1_TSR1_C
IPR037875 Bms1_N
IPR030387 G_Bms1/Tsr1_dom
IPR031483 AP1AR
IPR039761 Bms1/Tsr1
IPR027417 P-loop_NTPase
IPR012948 AARP2CN
IPR001714 Pept_M24_MAP
IPR002467 Pept_M24A_MAP1
IPR000994 Pept_M24
IPR036005 Creatinase/aminopeptidase-like
IPR007235 Glyco_trans_28_C
IPR007034 BMS1_TSR1_C
IPR037875 Bms1_N
IPR030387 G_Bms1/Tsr1_dom
IPR031483 AP1AR
IPR039761 Bms1/Tsr1
IPR027417 P-loop_NTPase
IPR012948 AARP2CN
ProteinModelPortal
H9IYR0
A0A2A4K1K6
S4P7N6
A0A2H1VWB4
A0A194QBM5
A0A212EJW8
+ More
A0A088AK91 D6X4P7 A0A0L7QQE3 A0A0M8ZVR5 A0A2A3ECH8 A0A1Y1NMM5 A0A0C9RES9 A0A026W4M4 A0A0T6B8S3 F4WU86 A0A158NTW4 A0A195C854 A0A232F417 A0A151JQQ9 K7IR17 E9IH60 E2BLG2 A0A182PTT1 A0A182W2Z1 A0A310SLT2 A0A182JYB7 A0A182WVF8 A0A182R8C9 A0A182I937 A0A182M4K1 R4WK40 A0A182YEU7 A0A182QXY3 A0A154PEU5 A0A182NCK4 A0A2J7RI72 A0A182KXY0 Q7QAH2 A0A182GP12 A0A182VLA5 Q0IFY6 A0A084WR40 E0VK43 A0A182JMZ4 A0A182SEW5 A0A1B6GC21 A0A1B6HYE0 B0WEW3 A0A182TJU8 A0A069DTH1 A0A182F8E1 A0A0A9Y188 A0A0P4VR59 A0A2M4BRK8 R4G8A5 W5JJK1 A0A1Q3FE60 A0A1B6LG69 A0A1B6CLC8 A0A2M4AQQ9 A0A0K8SLU0 A0A224YNV7 A0A131YSJ1 A0A194QSB9 A0A131X9M3 A0A023GJG4 G3MNE0 A0A023FKV8 A0A1B0C8Y0 U4USJ6 A0A1L8E2P3 L7M8Z5 A0A0P5K4P4 A0A1Z5L5F0 A0A0P6ASA1 A0A2R5LM79 A0A0P5G1E8 A0A0P5T2W0 A0A1S3D0B2 A0A336L8V3 A0A1E1X561 A0A0K8RD90 A0A293MTK8 U5EZB0 A0A0C9SCA6 A0A1W4W2P6 E2AWT8 E9GAH7 A0A1A9WC32 B4PV57 A0A067QZU9 B4HH59 T1G1H4 Q9VC48 A0A210R4V1 B3M040 A0A1B0A8U0 T1IS65 A0A034WMP3
A0A088AK91 D6X4P7 A0A0L7QQE3 A0A0M8ZVR5 A0A2A3ECH8 A0A1Y1NMM5 A0A0C9RES9 A0A026W4M4 A0A0T6B8S3 F4WU86 A0A158NTW4 A0A195C854 A0A232F417 A0A151JQQ9 K7IR17 E9IH60 E2BLG2 A0A182PTT1 A0A182W2Z1 A0A310SLT2 A0A182JYB7 A0A182WVF8 A0A182R8C9 A0A182I937 A0A182M4K1 R4WK40 A0A182YEU7 A0A182QXY3 A0A154PEU5 A0A182NCK4 A0A2J7RI72 A0A182KXY0 Q7QAH2 A0A182GP12 A0A182VLA5 Q0IFY6 A0A084WR40 E0VK43 A0A182JMZ4 A0A182SEW5 A0A1B6GC21 A0A1B6HYE0 B0WEW3 A0A182TJU8 A0A069DTH1 A0A182F8E1 A0A0A9Y188 A0A0P4VR59 A0A2M4BRK8 R4G8A5 W5JJK1 A0A1Q3FE60 A0A1B6LG69 A0A1B6CLC8 A0A2M4AQQ9 A0A0K8SLU0 A0A224YNV7 A0A131YSJ1 A0A194QSB9 A0A131X9M3 A0A023GJG4 G3MNE0 A0A023FKV8 A0A1B0C8Y0 U4USJ6 A0A1L8E2P3 L7M8Z5 A0A0P5K4P4 A0A1Z5L5F0 A0A0P6ASA1 A0A2R5LM79 A0A0P5G1E8 A0A0P5T2W0 A0A1S3D0B2 A0A336L8V3 A0A1E1X561 A0A0K8RD90 A0A293MTK8 U5EZB0 A0A0C9SCA6 A0A1W4W2P6 E2AWT8 E9GAH7 A0A1A9WC32 B4PV57 A0A067QZU9 B4HH59 T1G1H4 Q9VC48 A0A210R4V1 B3M040 A0A1B0A8U0 T1IS65 A0A034WMP3
PDB
4IU6
E-value=5.42359e-156,
Score=1413
Ontologies
KEGG
GO
PANTHER
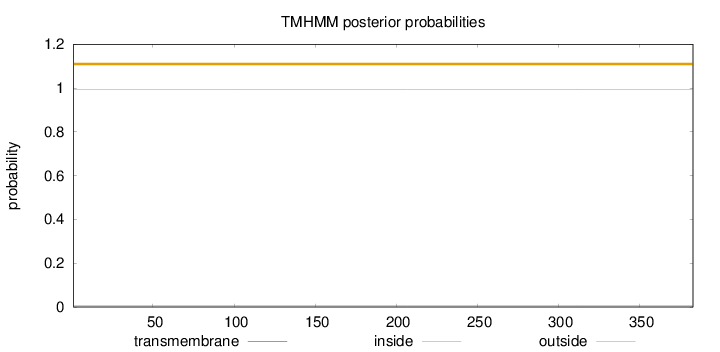
Topology
Length:
383
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00217
Exp number, first 60 AAs:
0.00217
Total prob of N-in:
0.00612
outside
1 - 383
Population Genetic Test Statistics
Pi
304.407119
Theta
216.675805
Tajima's D
1.274865
CLR
0
CSRT
0.733863306834658
Interpretation
Uncertain
