Gene
KWMTBOMO05317
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC100878911_[Megachile_rotundata]
Full name
Fatty acyl-CoA reductase
+ More
NAD-dependent protein deacylase
NAD-dependent protein deacylase
Alternative Name
Regulatory protein SIR2 homolog
Location in the cell
Mitochondrial Reliability : 2.17
Sequence
CDS
ATGACAGGATTCAGGAAGAGGAATCCTGAAATATCTGTTCGCATGTTCCAAAACCTTACTAGATCAAGAGCTGTAATATCCAAGGAGCATTTTAAAGAGGTACAGAGATTCTTGGATCAGACTCAACAAAAATACATCTTACAAGATCTTAGAAGGATCTTTAATACAGATGAAACTGCTTTTATTCTAAATCCGAAGGGTGGTAAGGTTTTAGCAAAAAAGGGGAACGAGAAAGTATATCAGCAAGTCAATCCGGATGAGAAAGAATGCCTATCAGTTCTTGTGACGGGTAGCGCATCTGGTGACCTCCTCCAACAGTTTTGTTTAAGTATAAAATAA
Protein
MTGFRKRNPEISVRMFQNLTRSRAVISKEHFKEVQRFLDQTQQKYILQDLRRIFNTDETAFILNPKGGKVLAKKGNEKVYQQVNPDEKECLSVLVTGSASGDLLQQFCLSIK
Summary
Description
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
NAD-dependent protein deacylase. Catalyzes the NAD-dependent hydrolysis of acyl groups from lysine residues.
NAD-dependent protein deacylase. Catalyzes the NAD-dependent hydrolysis of acyl groups from lysine residues.
Catalytic Activity
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
H2O + N(6)-acetyl-L-lysyl-[protein] + NAD(+) = 2''-O-acetyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
H2O + N(6)-acetyl-L-lysyl-[protein] + NAD(+) = 2''-O-acetyl-ADP-D-ribose + L-lysyl-[protein] + nicotinamide
Cofactor
Zn(2+)
Similarity
Belongs to the fatty acyl-CoA reductase family.
Belongs to the sirtuin family. Class II subfamily.
Belongs to the sirtuin family. Class II subfamily.
Uniprot
A0A224XMA5
A0A1Y1MDI2
H9JJ11
A0A0A9X621
A0A0A9X0L6
A0A0J7KF01
+ More
A0A1Y1KC33 A0A336KT24 A0A336L8I1 E9J8S3 A0A3L8E170 A0A1S4EH29 A0A3L8DK92 H9JW02 A0A0J7N979 A0A0A9W8Z7 J9K4C6 J9KY69 A0A0L7K2A4 A0A1B6G7C3 A0A3Q0JHK5 A0A151I654 A0A0L7K8J9 J9JM91 J9M4D1 A0A151J810 J9L8K0 H9JU07 J9KMK9 A0A2G8JN90 K7JEB0 X1X0E1 J9LC60 A0A0J7MZB4 A8E0Z6 J9K1I8 A0A1E1WTE0 A0A1B6MFI3 A0A2S2NKL5 A0A0J7K1X7 A0A182XPH4 X1XJF5 A0A2S2NUN9 J9L927 A0A2S2NH94 K7JEG9 W4YGX8 A0A2H1V3I1 A0A1S4EWX1 A0A0J7K2Z5 J9K668 A0A1Y1LYQ2 A0A1Y1LYU9 A0A1W7R625 A0A0P6IZY2 A0A2H1WY43 X1X791 X1WKJ5 A0A0L7L4B7 A0A182RZX0 X1WWX4 K7JP93 W4XWS8 A0A2S2NI89 A0A146NU82 A0A0N0PCP7 A0A2G8L1I0 A0A232EES1 X1XIJ7 A0A182PX43 A0A1B0DJ12 X1X3T4 K7JYS7 A0A2G8KHW0 A0A2C9KVQ1 A0A1X7SJH2 A0A2C9LKU7 K7JIQ4 J9JNH9 K7JHX5 J9LHM9 A0A1J1IVU2 A0A151J1R0 A0A2C9LK08 J9L1T1 A0A0C9Q4V7 A0A336MFM0 K7JGS0 A0A2C9KFK5 A0A2C9LW81 A0A1Y1K439 A0A2S2R564 A0A182I8H3
A0A1Y1KC33 A0A336KT24 A0A336L8I1 E9J8S3 A0A3L8E170 A0A1S4EH29 A0A3L8DK92 H9JW02 A0A0J7N979 A0A0A9W8Z7 J9K4C6 J9KY69 A0A0L7K2A4 A0A1B6G7C3 A0A3Q0JHK5 A0A151I654 A0A0L7K8J9 J9JM91 J9M4D1 A0A151J810 J9L8K0 H9JU07 J9KMK9 A0A2G8JN90 K7JEB0 X1X0E1 J9LC60 A0A0J7MZB4 A8E0Z6 J9K1I8 A0A1E1WTE0 A0A1B6MFI3 A0A2S2NKL5 A0A0J7K1X7 A0A182XPH4 X1XJF5 A0A2S2NUN9 J9L927 A0A2S2NH94 K7JEG9 W4YGX8 A0A2H1V3I1 A0A1S4EWX1 A0A0J7K2Z5 J9K668 A0A1Y1LYQ2 A0A1Y1LYU9 A0A1W7R625 A0A0P6IZY2 A0A2H1WY43 X1X791 X1WKJ5 A0A0L7L4B7 A0A182RZX0 X1WWX4 K7JP93 W4XWS8 A0A2S2NI89 A0A146NU82 A0A0N0PCP7 A0A2G8L1I0 A0A232EES1 X1XIJ7 A0A182PX43 A0A1B0DJ12 X1X3T4 K7JYS7 A0A2G8KHW0 A0A2C9KVQ1 A0A1X7SJH2 A0A2C9LKU7 K7JIQ4 J9JNH9 K7JHX5 J9LHM9 A0A1J1IVU2 A0A151J1R0 A0A2C9LK08 J9L1T1 A0A0C9Q4V7 A0A336MFM0 K7JGS0 A0A2C9KFK5 A0A2C9LW81 A0A1Y1K439 A0A2S2R564 A0A182I8H3
EC Number
1.2.1.84
3.5.1.-
3.5.1.-
Pubmed
EMBL
GFTR01007287
JAW09139.1
GEZM01036846
JAV82375.1
BABH01023137
GBHO01029341
+ More
JAG14263.1 GBHO01029337 JAG14267.1 LBMM01008491 KMQ88832.1 GEZM01087011 JAV58914.1 UFQS01000884 UFQT01000884 SSX07497.1 SSX27837.1 UFQS01002482 UFQT01002482 SSX14138.1 SSX33554.1 GL769049 EFZ10780.1 QOIP01000001 RLU26272.1 QOIP01000007 RLU20864.1 BABH01036510 LBMM01008012 KMQ89220.1 GBHO01039345 JAG04259.1 ABLF02013955 ABLF02007555 JTDY01015613 KOB51941.1 GECZ01011460 JAS58309.1 KQ978511 KYM93390.1 JTDY01010230 KOB59708.1 ABLF02017693 ABLF02017444 KQ979615 KYN20115.1 ABLF02006580 BABH01003862 ABLF02007625 ABLF02020143 MRZV01001531 PIK37236.1 AAZX01000647 AAZX01004403 AAZX01010750 AAZX01017323 ABLF02061385 ABLF02023847 LBMM01013145 KMQ85800.1 AM850132 CAO98966.1 ABLF02038792 GDQN01000745 JAT90309.1 GEBQ01005260 JAT34717.1 GGMR01005058 MBY17677.1 LBMM01016829 KMQ84299.1 ABLF02038785 GGMR01008133 MBY20752.1 ABLF02039566 GGMR01003952 MBY16571.1 AAZX01007240 AAZX01008809 AAZX01008867 AAZX01017227 AAZX01018662 AAZX01023390 AAZX01023918 AAZX01026511 AAGJ04065074 AAGJ04065075 ODYU01000509 SOQ35403.1 LBMM01015444 KMQ84803.1 ABLF02017438 ABLF02017439 GEZM01043830 JAV78744.1 GEZM01043833 JAV78742.1 GEHC01001021 JAV46624.1 GDUN01000556 JAN95363.1 ODYU01011524 SOQ57304.1 ABLF02064650 ABLF02025656 ABLF02025658 JTDY01003103 KOB70151.1 ABLF02017764 AAZX01014012 AAGJ04120197 GGMR01004250 MBY16869.1 GCES01151013 JAQ35309.1 KQ460474 KPJ14372.1 MRZV01000262 PIK54124.1 NNAY01005248 OXU16828.1 ABLF02016564 AJVK01063592 ABLF02004347 AAZX01009120 MRZV01000572 PIK47582.1 AAZX01014011 ABLF02024890 AAZX01012377 ABLF02010575 CVRI01000058 CRL02657.1 KQ980507 KYN15781.1 ABLF02017442 ABLF02017446 GBYB01009113 JAG78880.1 UFQS01000961 UFQS01001389 UFQT01000961 UFQT01001389 SSX07977.1 SSX10646.1 SSX28211.1 GEZM01095923 GEZM01095922 JAV55258.1 GGMS01015966 MBY85169.1 APCN01007703
JAG14263.1 GBHO01029337 JAG14267.1 LBMM01008491 KMQ88832.1 GEZM01087011 JAV58914.1 UFQS01000884 UFQT01000884 SSX07497.1 SSX27837.1 UFQS01002482 UFQT01002482 SSX14138.1 SSX33554.1 GL769049 EFZ10780.1 QOIP01000001 RLU26272.1 QOIP01000007 RLU20864.1 BABH01036510 LBMM01008012 KMQ89220.1 GBHO01039345 JAG04259.1 ABLF02013955 ABLF02007555 JTDY01015613 KOB51941.1 GECZ01011460 JAS58309.1 KQ978511 KYM93390.1 JTDY01010230 KOB59708.1 ABLF02017693 ABLF02017444 KQ979615 KYN20115.1 ABLF02006580 BABH01003862 ABLF02007625 ABLF02020143 MRZV01001531 PIK37236.1 AAZX01000647 AAZX01004403 AAZX01010750 AAZX01017323 ABLF02061385 ABLF02023847 LBMM01013145 KMQ85800.1 AM850132 CAO98966.1 ABLF02038792 GDQN01000745 JAT90309.1 GEBQ01005260 JAT34717.1 GGMR01005058 MBY17677.1 LBMM01016829 KMQ84299.1 ABLF02038785 GGMR01008133 MBY20752.1 ABLF02039566 GGMR01003952 MBY16571.1 AAZX01007240 AAZX01008809 AAZX01008867 AAZX01017227 AAZX01018662 AAZX01023390 AAZX01023918 AAZX01026511 AAGJ04065074 AAGJ04065075 ODYU01000509 SOQ35403.1 LBMM01015444 KMQ84803.1 ABLF02017438 ABLF02017439 GEZM01043830 JAV78744.1 GEZM01043833 JAV78742.1 GEHC01001021 JAV46624.1 GDUN01000556 JAN95363.1 ODYU01011524 SOQ57304.1 ABLF02064650 ABLF02025656 ABLF02025658 JTDY01003103 KOB70151.1 ABLF02017764 AAZX01014012 AAGJ04120197 GGMR01004250 MBY16869.1 GCES01151013 JAQ35309.1 KQ460474 KPJ14372.1 MRZV01000262 PIK54124.1 NNAY01005248 OXU16828.1 ABLF02016564 AJVK01063592 ABLF02004347 AAZX01009120 MRZV01000572 PIK47582.1 AAZX01014011 ABLF02024890 AAZX01012377 ABLF02010575 CVRI01000058 CRL02657.1 KQ980507 KYN15781.1 ABLF02017442 ABLF02017446 GBYB01009113 JAG78880.1 UFQS01000961 UFQS01001389 UFQT01000961 UFQT01001389 SSX07977.1 SSX10646.1 SSX28211.1 GEZM01095923 GEZM01095922 JAV55258.1 GGMS01015966 MBY85169.1 APCN01007703
Proteomes
Pfam
Interpro
IPR009057
Homeobox-like_sf
+ More
IPR007889 HTH_Psq
IPR006600 HTH_CenpB_DNA-bd_dom
IPR004875 DDE_SF_endonuclease_dom
IPR005312 DUF1759
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR029196 DUF4588
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR031961 DUF4780
IPR026055 FAR
IPR033640 FAR_C
IPR013120 Male_sterile_NAD-bd
IPR036291 NAD(P)-bd_dom_sf
IPR002219 PE/DAG-bd
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR026590 Ssirtuin_cat_dom
IPR026587 Sirtuin_class_II
IPR026591 Sirtuin_cat_small_dom_sf
IPR003000 Sirtuin
IPR007889 HTH_Psq
IPR006600 HTH_CenpB_DNA-bd_dom
IPR004875 DDE_SF_endonuclease_dom
IPR005312 DUF1759
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR029196 DUF4588
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR031961 DUF4780
IPR026055 FAR
IPR033640 FAR_C
IPR013120 Male_sterile_NAD-bd
IPR036291 NAD(P)-bd_dom_sf
IPR002219 PE/DAG-bd
IPR029035 DHS-like_NAD/FAD-binding_dom
IPR026590 Ssirtuin_cat_dom
IPR026587 Sirtuin_class_II
IPR026591 Sirtuin_cat_small_dom_sf
IPR003000 Sirtuin
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A224XMA5
A0A1Y1MDI2
H9JJ11
A0A0A9X621
A0A0A9X0L6
A0A0J7KF01
+ More
A0A1Y1KC33 A0A336KT24 A0A336L8I1 E9J8S3 A0A3L8E170 A0A1S4EH29 A0A3L8DK92 H9JW02 A0A0J7N979 A0A0A9W8Z7 J9K4C6 J9KY69 A0A0L7K2A4 A0A1B6G7C3 A0A3Q0JHK5 A0A151I654 A0A0L7K8J9 J9JM91 J9M4D1 A0A151J810 J9L8K0 H9JU07 J9KMK9 A0A2G8JN90 K7JEB0 X1X0E1 J9LC60 A0A0J7MZB4 A8E0Z6 J9K1I8 A0A1E1WTE0 A0A1B6MFI3 A0A2S2NKL5 A0A0J7K1X7 A0A182XPH4 X1XJF5 A0A2S2NUN9 J9L927 A0A2S2NH94 K7JEG9 W4YGX8 A0A2H1V3I1 A0A1S4EWX1 A0A0J7K2Z5 J9K668 A0A1Y1LYQ2 A0A1Y1LYU9 A0A1W7R625 A0A0P6IZY2 A0A2H1WY43 X1X791 X1WKJ5 A0A0L7L4B7 A0A182RZX0 X1WWX4 K7JP93 W4XWS8 A0A2S2NI89 A0A146NU82 A0A0N0PCP7 A0A2G8L1I0 A0A232EES1 X1XIJ7 A0A182PX43 A0A1B0DJ12 X1X3T4 K7JYS7 A0A2G8KHW0 A0A2C9KVQ1 A0A1X7SJH2 A0A2C9LKU7 K7JIQ4 J9JNH9 K7JHX5 J9LHM9 A0A1J1IVU2 A0A151J1R0 A0A2C9LK08 J9L1T1 A0A0C9Q4V7 A0A336MFM0 K7JGS0 A0A2C9KFK5 A0A2C9LW81 A0A1Y1K439 A0A2S2R564 A0A182I8H3
A0A1Y1KC33 A0A336KT24 A0A336L8I1 E9J8S3 A0A3L8E170 A0A1S4EH29 A0A3L8DK92 H9JW02 A0A0J7N979 A0A0A9W8Z7 J9K4C6 J9KY69 A0A0L7K2A4 A0A1B6G7C3 A0A3Q0JHK5 A0A151I654 A0A0L7K8J9 J9JM91 J9M4D1 A0A151J810 J9L8K0 H9JU07 J9KMK9 A0A2G8JN90 K7JEB0 X1X0E1 J9LC60 A0A0J7MZB4 A8E0Z6 J9K1I8 A0A1E1WTE0 A0A1B6MFI3 A0A2S2NKL5 A0A0J7K1X7 A0A182XPH4 X1XJF5 A0A2S2NUN9 J9L927 A0A2S2NH94 K7JEG9 W4YGX8 A0A2H1V3I1 A0A1S4EWX1 A0A0J7K2Z5 J9K668 A0A1Y1LYQ2 A0A1Y1LYU9 A0A1W7R625 A0A0P6IZY2 A0A2H1WY43 X1X791 X1WKJ5 A0A0L7L4B7 A0A182RZX0 X1WWX4 K7JP93 W4XWS8 A0A2S2NI89 A0A146NU82 A0A0N0PCP7 A0A2G8L1I0 A0A232EES1 X1XIJ7 A0A182PX43 A0A1B0DJ12 X1X3T4 K7JYS7 A0A2G8KHW0 A0A2C9KVQ1 A0A1X7SJH2 A0A2C9LKU7 K7JIQ4 J9JNH9 K7JHX5 J9LHM9 A0A1J1IVU2 A0A151J1R0 A0A2C9LK08 J9L1T1 A0A0C9Q4V7 A0A336MFM0 K7JGS0 A0A2C9KFK5 A0A2C9LW81 A0A1Y1K439 A0A2S2R564 A0A182I8H3
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
Mitochondrion matrix
Mitochondrion matrix
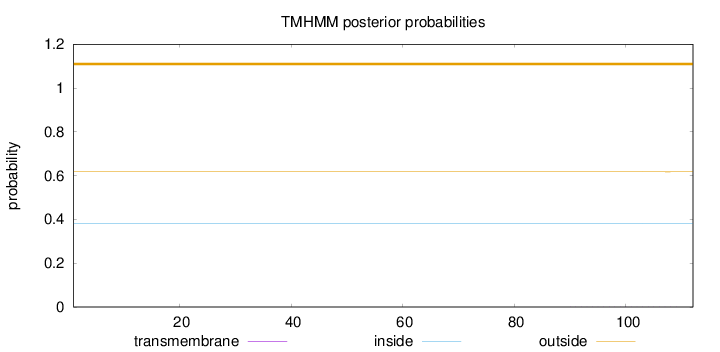
Length:
112
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00365
Exp number, first 60 AAs:
0
Total prob of N-in:
0.38267
outside
1 - 112
Population Genetic Test Statistics
Pi
330.496342
Theta
214.040512
Tajima's D
1.762546
CLR
0.336573
CSRT
0.844207789610519
Interpretation
Uncertain
