Gene
KWMTBOMO05301
Pre Gene Modal
BGIBMGA002400
Annotation
PREDICTED:_calcium-activated_potassium_channel_slowpoke_[Bombyx_mori]
Full name
Calcium-activated potassium channel slowpoke
Alternative Name
BK channel
Maxi K channel
Maxi K channel
Location in the cell
PlasmaMembrane Reliability : 3.358
Sequence
CDS
ATGGTCTACTTTTTTATACGATTTATAGCGGCTAGTGACAAACTATGGTTCATGCTCGAGATGTATTCGTTTGTAGATTATTTCACGATACCCCCATCATTTGTATCGATATACTTGGACAGAACGTGGATAGGTCTGAGATTTCTCAGGGCTCTGCGTTTAATGACCGTGCCTGATATTTTGCAATACCTCAATATATTAAAAACGTCAAGCTCAATCCGCTTAGCGCAATTAGTTTCTATATTTATATCAGTGTGGTTAACAGCCGCCGGTATTATTCATTTGCTGGAAAACTCAGGGGATCCGTTGGAATTCGCTAACTCACAGAAGCTCTCATACTGGACATGCGTGTATTTCCTTATAGTGACGATGTCCACAGTAGGTTACGGTGATGTGTTCTGTCACACTGTACTAGGCAGAACCTTTTTGGTATTTTTTCTCCTCGTTGGTTTGGCGATATTTGCCAGTTGTATCCCAGAGATAATTGACTTAATCGGCACTAGACCCAAGTACGGCGGCACCCTCAAGAATGAGCGGGGACGCAGGCACATTGTTGTATGTGGCCATATAACATACGAATCAGTGAGTCATTTTTTAAAAGACTTCCTACACGAAGATCGAGAGGATGTGGATGTTGAGGTGGTTTTTTTACACAGGAAACCGCCCGACCTGGAGCTCGAAGGCCTGTTCAAGAGACACTTTACCACTGTGGAATTCTTTCAAGGCACCATTATGAATCCAATCGACCTGCAAAGGGTAAAAGTCCATGAAGCAGATGCTTGTCTGGTGCTAGCCAACAAATACTGCCAGGATCCGGACGCTGAGGACGCAGCCAATATCATGAGGGTCATCTCTATCAAGAACTACTCCGACGATATCAGGGTCATCATACAGCTCATGCAGTACCATAATAAGGCCTATCTGTTAAACATTCCATCATGGGATTGGAAGCAAGGAGACGACGTGATTTGCTTGGCGGAACTTAAACTCGGCTTCATCGCCCAGAGCTGCCTTGCGCCTGGCTTCTCGACCATGATGGCTAACCTCTTCGCCATGCGGAGCTTCAAGACGTCCCAAGACATGGCTATATGGACTAATGACTACATGCGAGGTACCGGGATGGAGATGTACACAGAGACCCTTAGTCCTACGTTTATCGGGATGCCTTTCATTGAAGCAGCCGAGTTATGCTTTACGAAACTCAAGCTTCTTCTATTAGCAATAGAGATCAAGGGAGAAGAGGGAGCCGATAGCAAAATATCGATAAACCCTCGCAACGCGAAAATCCATGCCAATACCCAAGGGTTCTTCATAGCACAGTCGGCTGATGAAGTCAAAAGAGCGTGGTTTTATTGTAAGGCCTGCCACGAGGACATCAAAGACGAAACACTCATCAAGAAGTGCAAATGCAAAAACTATGTCGGAATGATGATGATGCAGACTGGGATGGTGAAACAAAGTCTTAATACGTACCGTGCTCTCGATGATGATCACCATCCTCCCCCCACCTTCACGCCGCCAGAGTTGCCCAAGAAGGTCCATGTTCGAGGCGAAATCGCCCGCGAAAGGGGGGAAGATACAAACTTAATAAATCGCAACCATCGGAACGCGCCGACGGCGATGCTGTCTCCAATGGCGAACCAGCTGGTGAACACGACGACGACTCGGCAAGTCAACAAAGTAAAGCCGAACGTCAATCGAGTACCGCCCGATACGAATCCTCAAGATCAAGACACGAATTACCAGGCCTATCACCTTGCCTACGAAGTGAAAAAGCTCATGCCTACTAGCAGAAGCAGTGGGGGTAATCAGAACAGCAATGGAGTTAGTCTGCCTGCTGGATTGGCAGACGACCAGTCGAAAGACTTCGACTTTGAAAAGACTGAAATGAAATACGACTCAACAGGCATGTTCCATTGGAGCCCTGCCAGGAATCTTGAAGATTGTATATTAGACAGAAACCAAGCGGCTATGACGGTTCTAAATGGTCACGTTGTCGTGTGTCTATTCGCGGATCCAGACTCGCCACTGATCGGCTTGAGGAATCTCGTGATGCCGCTAAGAGCCTCCAATTTCCACTACCACGAGTTGAAACACGTGGTTATCGTAGGAAGCGTCGACTACATACGAAGAGAATGGAAAATGTTGCAGAATCTTCCGAAGATATCTGTATTGAATGGTTCACCGCTAAGCCGGGCCGACTTGCGTGCAGTAAACGTGAACCTATGCGACATGTGCTGTATCCTGTCAGCGAAGGTGCCATCGAACGATGACCCTACACTGGCGGACAAGGAAGCTATCTTGGCTTCGTTGAACATCAAGGCGATGACGTTCGACGACACGATAGGAGTGCTCAGCGCCGGCGCGGGGGCCAACGGCAGCAACGCGCAGCCGCTGCCGGGCCAGCCCCCCGCGCCCGTGCTGCTACAGAGGAGGGGATCCGTATACGGTGCTAATGTACCTATGATTACAGAGCTGGTGAACGATAGCAACGTGCAGTTCTTAGACCAGGACGACGACGACGACCCCGACACGGAGCTGTACTTGACGCAACCGTTTGCCTGCGGCACTGCGTTTGCCGTCAGCGTACTCGACTCTCTCATGTCCACTACGTACTTCAACCAAAACGCGTTGACACTGATCCGTTCGCTGATAACGGGCGGCGCGACCCCCGAGCTGGAGCTGATCCTCGCCGAAGGTGCCGGGCTCCGCGGCGGGTACTCCACGCCCGAGAGTCTGGCCAACAGGTCTCCGCACTACTCACTAAACAAATACCAAACTCGATGA
Protein
MVYFFIRFIAASDKLWFMLEMYSFVDYFTIPPSFVSIYLDRTWIGLRFLRALRLMTVPDILQYLNILKTSSSIRLAQLVSIFISVWLTAAGIIHLLENSGDPLEFANSQKLSYWTCVYFLIVTMSTVGYGDVFCHTVLGRTFLVFFLLVGLAIFASCIPEIIDLIGTRPKYGGTLKNERGRRHIVVCGHITYESVSHFLKDFLHEDREDVDVEVVFLHRKPPDLELEGLFKRHFTTVEFFQGTIMNPIDLQRVKVHEADACLVLANKYCQDPDAEDAANIMRVISIKNYSDDIRVIIQLMQYHNKAYLLNIPSWDWKQGDDVICLAELKLGFIAQSCLAPGFSTMMANLFAMRSFKTSQDMAIWTNDYMRGTGMEMYTETLSPTFIGMPFIEAAELCFTKLKLLLLAIEIKGEEGADSKISINPRNAKIHANTQGFFIAQSADEVKRAWFYCKACHEDIKDETLIKKCKCKNYVGMMMMQTGMVKQSLNTYRALDDDHHPPPTFTPPELPKKVHVRGEIARERGEDTNLINRNHRNAPTAMLSPMANQLVNTTTTRQVNKVKPNVNRVPPDTNPQDQDTNYQAYHLAYEVKKLMPTSRSSGGNQNSNGVSLPAGLADDQSKDFDFEKTEMKYDSTGMFHWSPARNLEDCILDRNQAAMTVLNGHVVVCLFADPDSPLIGLRNLVMPLRASNFHYHELKHVVIVGSVDYIRREWKMLQNLPKISVLNGSPLSRADLRAVNVNLCDMCCILSAKVPSNDDPTLADKEAILASLNIKAMTFDDTIGVLSAGAGANGSNAQPLPGQPPAPVLLQRRGSVYGANVPMITELVNDSNVQFLDQDDDDDPDTELYLTQPFACGTAFAVSVLDSLMSTTYFNQNALTLIRSLITGGATPELELILAEGAGLRGGYSTPESLANRSPHYSLNKYQTR
Summary
Description
Potassium channel activated by both membrane depolarization or increase in cytosolic Ca(2+) that mediates export of K(+). Its activation dampens the excitatory events that elevate the cytosolic Ca(2+) concentration and/or depolarize the cell membrane. It therefore contributes to repolarization of the membrane potential. Kinetics are determined by alternative splicing, phosphorylation status and its combination interaction with Slob and 14-3-3-zeta. While the interaction with Slob1 alone increases its activity, its interaction with both Slob1 and 14-3-3-zeta decreases its activity.
Subunit
Homotetramer; which constitutes the calcium-activated potassium channel (By similarity). Interacts with Slip1. Interacts with Slob, and, indirectly with 14-3-3-zeta via its interaction with Slob. Interacts with Pka-C1 and Src kinases, which can bind simultaneously to it.
Similarity
Belongs to the potassium channel family. Calcium-activated (TC 1.A.1.3) subfamily. Slo sub-subfamily.
Keywords
Alternative splicing
Calcium
Complete proteome
Ion channel
Ion transport
Membrane
Phosphoprotein
Potassium
Potassium channel
Potassium transport
Reference proteome
Transmembrane
Transmembrane helix
Transport
Voltage-gated channel
Feature
chain Calcium-activated potassium channel slowpoke
splice variant In isoform 6, isoform B and isoform C.
splice variant In isoform 6, isoform B and isoform C.
Uniprot
A0A3S2L4Y1
A0A2A4J791
A0A1I8NVX3
Q4VS08
A0A1I8NVV0
A0A1I8NVY2
+ More
A0A1I8NVW4 A0A0R3NEJ5 A0A1W4VQ27 A0A0Q9X5Q4 A0A1W4W270 A0A0K8V1I6 A0A1W4W2R8 A0A2J7Q2E0 A0A0Q9X757 A0A146M5Z5 A0A1W4VPS9 A0A1I8NVW5 A0A1I8NVY4 A0A1I8NVX6 A0A1I8NVX4 A0A1W4VQ34 A0A034VBF9 A0A0Q9WZL0 A0A151WZ87 B4JEJ3 A0A1I8NVV9 A0A1L8DEY2 A0A0R3NEL9 A0A034VC34 A0A1B6JC67 A0A1I8NCH7 A0A1B6J7W9 A0A0R1E5N0 A0A1Q3EUQ3 A0A0Q9WXN9 A0A1W4W250 Q03720-2 A0A1W4W1B4 A0A1I8NVU4 A0A1W4W2S7 A0A0R3NK01 A0A0R1E5Q1 B3M043 Q03720-3 I5AMY0 D6X3R4 A0A1W4W2T4 A0A0Q9WZM6 A0A1I8NVW2 A0A1I8NVV6 A0A1W4VQ22 B4K514 A0A1W4W1B9 A0A0Q9WXN5 A0A0R1EAG5 A0A0R3NKF6 A0A195F4I3 A0A1I8NVW6 A0A1W4W274 A0A0Q9WPI2 A0A1I8NVW0 A0A1W4VPT4 A0A0Q9WYF8 A0A0Q9WXS2 A0A0R3NF49 Q03720-4 A0A1I8NVX8 A0A1I8NCH2 A0A1I8NVU8 B4LW34 A0A1I8NCG3 A0A1W4VQ40 A0A1I8NCG2 A0A0R3NK40 A0A0P5ESN0 A0A0Q9X9R3 A0A1I8NVY5 A0A1J1HU24 A0A0N8P1C0 A0A0P8YDD8 A0A0R3NIX5 Q03720-16 A0A1W4W2U8 A0A0R1EB14 A0A0P8XXN1 A0A1I8NCH0 A0A0P8XXU9 A0A0Q9WTD6 A0A1W4VPS4 A0A0R3NK22 A0A0Q9X1Q9 A0A0Q9WXL9 B4PVR0 A0A1W4VQ48 A0A0B4KHT2 A0A0R1E5P6 A0A3B0JLT8 A0A0R1E5R2 A0A0Q9WDW2
A0A1I8NVW4 A0A0R3NEJ5 A0A1W4VQ27 A0A0Q9X5Q4 A0A1W4W270 A0A0K8V1I6 A0A1W4W2R8 A0A2J7Q2E0 A0A0Q9X757 A0A146M5Z5 A0A1W4VPS9 A0A1I8NVW5 A0A1I8NVY4 A0A1I8NVX6 A0A1I8NVX4 A0A1W4VQ34 A0A034VBF9 A0A0Q9WZL0 A0A151WZ87 B4JEJ3 A0A1I8NVV9 A0A1L8DEY2 A0A0R3NEL9 A0A034VC34 A0A1B6JC67 A0A1I8NCH7 A0A1B6J7W9 A0A0R1E5N0 A0A1Q3EUQ3 A0A0Q9WXN9 A0A1W4W250 Q03720-2 A0A1W4W1B4 A0A1I8NVU4 A0A1W4W2S7 A0A0R3NK01 A0A0R1E5Q1 B3M043 Q03720-3 I5AMY0 D6X3R4 A0A1W4W2T4 A0A0Q9WZM6 A0A1I8NVW2 A0A1I8NVV6 A0A1W4VQ22 B4K514 A0A1W4W1B9 A0A0Q9WXN5 A0A0R1EAG5 A0A0R3NKF6 A0A195F4I3 A0A1I8NVW6 A0A1W4W274 A0A0Q9WPI2 A0A1I8NVW0 A0A1W4VPT4 A0A0Q9WYF8 A0A0Q9WXS2 A0A0R3NF49 Q03720-4 A0A1I8NVX8 A0A1I8NCH2 A0A1I8NVU8 B4LW34 A0A1I8NCG3 A0A1W4VQ40 A0A1I8NCG2 A0A0R3NK40 A0A0P5ESN0 A0A0Q9X9R3 A0A1I8NVY5 A0A1J1HU24 A0A0N8P1C0 A0A0P8YDD8 A0A0R3NIX5 Q03720-16 A0A1W4W2U8 A0A0R1EB14 A0A0P8XXN1 A0A1I8NCH0 A0A0P8XXU9 A0A0Q9WTD6 A0A1W4VPS4 A0A0R3NK22 A0A0Q9X1Q9 A0A0Q9WXL9 B4PVR0 A0A1W4VQ48 A0A0B4KHT2 A0A0R1E5P6 A0A3B0JLT8 A0A0R1E5R2 A0A0Q9WDW2
Pubmed
EMBL
RSAL01000148
RVE45825.1
NWSH01002925
PCG67283.1
AY644784
AAT44358.1
+ More
CM000070 KRS99607.1 CH933806 KRG00756.1 GDHF01019849 JAI32465.1 NEVH01019371 PNF22750.1 KRG00770.1 GDHC01004432 JAQ14197.1 GAKP01019842 JAC39110.1 KRG00761.1 KQ982649 KYQ53198.1 CH916369 EDV93124.1 GFDF01009086 JAV04998.1 KRS99617.1 GAKP01019839 JAC39113.1 GECU01011143 JAS96563.1 GECU01012448 JAS95258.1 CM000160 KRK04534.1 GFDL01016010 JAV19035.1 KRG00784.1 M69053 M96840 AE014297 BT004876 U40315 KRS99604.1 KRK04520.1 CH902617 EDV42000.2 EIM52315.2 KQ971377 EEZ97457.2 KRG00777.1 EDW13985.2 KRG00774.1 KRK04545.1 KRS99618.1 KQ981805 KYN35490.1 CH940650 KRF82788.1 KRG00757.1 KRG00779.1 KRS99625.1 EDW66539.2 KRS99628.1 GDIQ01265070 JAJ86654.1 KRG00776.1 CVRI01000021 CRK91501.1 KPU79500.1 KPU79497.1 KRS99614.1 KRK04522.1 KPU79493.1 KPU79482.1 CH964232 KRF99477.1 KRS99620.1 KRF99475.1 KRG00783.1 EDW98909.2 AGB96304.1 KRK04528.1 OUUW01000005 SPP81292.1 KRK04540.1 KRF82793.1
CM000070 KRS99607.1 CH933806 KRG00756.1 GDHF01019849 JAI32465.1 NEVH01019371 PNF22750.1 KRG00770.1 GDHC01004432 JAQ14197.1 GAKP01019842 JAC39110.1 KRG00761.1 KQ982649 KYQ53198.1 CH916369 EDV93124.1 GFDF01009086 JAV04998.1 KRS99617.1 GAKP01019839 JAC39113.1 GECU01011143 JAS96563.1 GECU01012448 JAS95258.1 CM000160 KRK04534.1 GFDL01016010 JAV19035.1 KRG00784.1 M69053 M96840 AE014297 BT004876 U40315 KRS99604.1 KRK04520.1 CH902617 EDV42000.2 EIM52315.2 KQ971377 EEZ97457.2 KRG00777.1 EDW13985.2 KRG00774.1 KRK04545.1 KRS99618.1 KQ981805 KYN35490.1 CH940650 KRF82788.1 KRG00757.1 KRG00779.1 KRS99625.1 EDW66539.2 KRS99628.1 GDIQ01265070 JAJ86654.1 KRG00776.1 CVRI01000021 CRK91501.1 KPU79500.1 KPU79497.1 KRS99614.1 KRK04522.1 KPU79493.1 KPU79482.1 CH964232 KRF99477.1 KRS99620.1 KRF99475.1 KRG00783.1 EDW98909.2 AGB96304.1 KRK04528.1 OUUW01000005 SPP81292.1 KRK04540.1 KRF82793.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF51735
SSF51735
Gene 3D
ProteinModelPortal
A0A3S2L4Y1
A0A2A4J791
A0A1I8NVX3
Q4VS08
A0A1I8NVV0
A0A1I8NVY2
+ More
A0A1I8NVW4 A0A0R3NEJ5 A0A1W4VQ27 A0A0Q9X5Q4 A0A1W4W270 A0A0K8V1I6 A0A1W4W2R8 A0A2J7Q2E0 A0A0Q9X757 A0A146M5Z5 A0A1W4VPS9 A0A1I8NVW5 A0A1I8NVY4 A0A1I8NVX6 A0A1I8NVX4 A0A1W4VQ34 A0A034VBF9 A0A0Q9WZL0 A0A151WZ87 B4JEJ3 A0A1I8NVV9 A0A1L8DEY2 A0A0R3NEL9 A0A034VC34 A0A1B6JC67 A0A1I8NCH7 A0A1B6J7W9 A0A0R1E5N0 A0A1Q3EUQ3 A0A0Q9WXN9 A0A1W4W250 Q03720-2 A0A1W4W1B4 A0A1I8NVU4 A0A1W4W2S7 A0A0R3NK01 A0A0R1E5Q1 B3M043 Q03720-3 I5AMY0 D6X3R4 A0A1W4W2T4 A0A0Q9WZM6 A0A1I8NVW2 A0A1I8NVV6 A0A1W4VQ22 B4K514 A0A1W4W1B9 A0A0Q9WXN5 A0A0R1EAG5 A0A0R3NKF6 A0A195F4I3 A0A1I8NVW6 A0A1W4W274 A0A0Q9WPI2 A0A1I8NVW0 A0A1W4VPT4 A0A0Q9WYF8 A0A0Q9WXS2 A0A0R3NF49 Q03720-4 A0A1I8NVX8 A0A1I8NCH2 A0A1I8NVU8 B4LW34 A0A1I8NCG3 A0A1W4VQ40 A0A1I8NCG2 A0A0R3NK40 A0A0P5ESN0 A0A0Q9X9R3 A0A1I8NVY5 A0A1J1HU24 A0A0N8P1C0 A0A0P8YDD8 A0A0R3NIX5 Q03720-16 A0A1W4W2U8 A0A0R1EB14 A0A0P8XXN1 A0A1I8NCH0 A0A0P8XXU9 A0A0Q9WTD6 A0A1W4VPS4 A0A0R3NK22 A0A0Q9X1Q9 A0A0Q9WXL9 B4PVR0 A0A1W4VQ48 A0A0B4KHT2 A0A0R1E5P6 A0A3B0JLT8 A0A0R1E5R2 A0A0Q9WDW2
A0A1I8NVW4 A0A0R3NEJ5 A0A1W4VQ27 A0A0Q9X5Q4 A0A1W4W270 A0A0K8V1I6 A0A1W4W2R8 A0A2J7Q2E0 A0A0Q9X757 A0A146M5Z5 A0A1W4VPS9 A0A1I8NVW5 A0A1I8NVY4 A0A1I8NVX6 A0A1I8NVX4 A0A1W4VQ34 A0A034VBF9 A0A0Q9WZL0 A0A151WZ87 B4JEJ3 A0A1I8NVV9 A0A1L8DEY2 A0A0R3NEL9 A0A034VC34 A0A1B6JC67 A0A1I8NCH7 A0A1B6J7W9 A0A0R1E5N0 A0A1Q3EUQ3 A0A0Q9WXN9 A0A1W4W250 Q03720-2 A0A1W4W1B4 A0A1I8NVU4 A0A1W4W2S7 A0A0R3NK01 A0A0R1E5Q1 B3M043 Q03720-3 I5AMY0 D6X3R4 A0A1W4W2T4 A0A0Q9WZM6 A0A1I8NVW2 A0A1I8NVV6 A0A1W4VQ22 B4K514 A0A1W4W1B9 A0A0Q9WXN5 A0A0R1EAG5 A0A0R3NKF6 A0A195F4I3 A0A1I8NVW6 A0A1W4W274 A0A0Q9WPI2 A0A1I8NVW0 A0A1W4VPT4 A0A0Q9WYF8 A0A0Q9WXS2 A0A0R3NF49 Q03720-4 A0A1I8NVX8 A0A1I8NCH2 A0A1I8NVU8 B4LW34 A0A1I8NCG3 A0A1W4VQ40 A0A1I8NCG2 A0A0R3NK40 A0A0P5ESN0 A0A0Q9X9R3 A0A1I8NVY5 A0A1J1HU24 A0A0N8P1C0 A0A0P8YDD8 A0A0R3NIX5 Q03720-16 A0A1W4W2U8 A0A0R1EB14 A0A0P8XXN1 A0A1I8NCH0 A0A0P8XXU9 A0A0Q9WTD6 A0A1W4VPS4 A0A0R3NK22 A0A0Q9X1Q9 A0A0Q9WXL9 B4PVR0 A0A1W4VQ48 A0A0B4KHT2 A0A0R1E5P6 A0A3B0JLT8 A0A0R1E5R2 A0A0Q9WDW2
PDB
5TJI
E-value=0,
Score=1762
Ontologies
GO
PANTHER
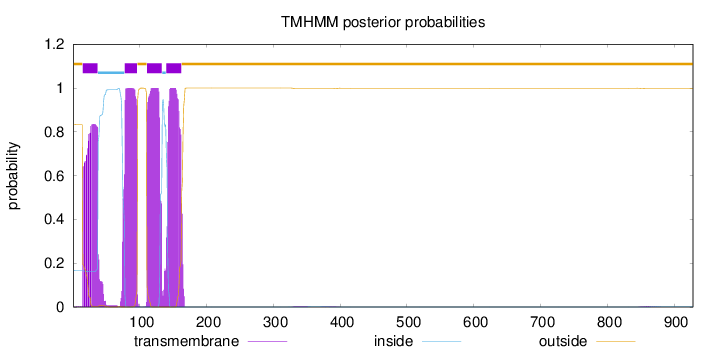
Topology
Subcellular location
Membrane
Length:
928
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
83.6358200000003
Exp number, first 60 AAs:
19.05992
Total prob of N-in:
0.16687
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 77
TMhelix
78 - 96
outside
97 - 110
TMhelix
111 - 133
inside
134 - 139
TMhelix
140 - 162
outside
163 - 928
Population Genetic Test Statistics
Pi
265.52197
Theta
187.839653
Tajima's D
1.209871
CLR
0.404745
CSRT
0.72046397680116
Interpretation
Uncertain
