Gene
KWMTBOMO05297 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002509
Annotation
PREDICTED:_tropomodulin_isoform_X5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.238
Sequence
CDS
ATGATGAATCAGGACCAGTATCACGCGTCGCTTCTGAACAAGGGCCAGCCCGTCGGACTCGGATGGGACGGCATCACCAAAGCCACTAAACCCAAGGTATTCCCAATGGACCCGCCCAACGACACGGACCCGGATGAAACGATCGCTCGAGTGCAACAAAATGACCAGAAGCTGACTGATCTCAATTGGAATAATATTAAGAACATAAGCGACGAGAAGTTCGAGAAGCTGTTCGAAGCGCTAAAGAGCAACAGTCACGTGGAGGTGCTGTCGCTGGTGAACGTGGGGCTGACGGACCGCACGGCGGGCGCGCTGGCGGACGCGCTGCGGCACAACACGGCGCTGCGCGTGCTCAGCGTCGAGACCAACTTCATCTCGCCGCCCGCGCTCGTGCAGCTCGTGCGCGCGCTGCTGCACACCAACACCGTCGAGGAGTTCCGGGCCTCCAACCAGAGATCGCAAGTTCTCGGTAACAAGATCGAGATGGAGATAACTCAGCTCGTCGAACAGAATCCGACCCTGCTGCGTCTCGGGCTGCACCTCGAGTACAGCGACGCGAGGCACAGGGTCGCGTCGCATCTCCAACGAAACATCGATAGGATACGCCAGCAGCGCCGGAGCCAGAATAGCGCTTAG
Protein
MMNQDQYHASLLNKGQPVGLGWDGITKATKPKVFPMDPPNDTDPDETIARVQQNDQKLTDLNWNNIKNISDEKFEKLFEALKSNSHVEVLSLVNVGLTDRTAGALADALRHNTALRVLSVETNFISPPALVQLVRALLHTNTVEEFRASNQRSQVLGNKIEMEITQLVEQNPTLLRLGLHLEYSDARHRVASHLQRNIDRIRQQRRSQNSA
Summary
Uniprot
A0A2H1VIY4
A0A194QBL0
A0A212ET25
A0A232FE74
A0A0T6BB80
A0A2S2PZ53
+ More
A0A139WKS9 A0A195EWW9 A0A158NB57 A0A139WL15 E2BYX3 A0A195D6L2 F4WXV5 A0A195BSR3 A0A151X0G5 A0A0L7QTH0 E2ARL7 A0A3S2LWV0 A0A2A3EMU1 A0A1B6HBC5 V5GSJ1 A0A088A0V7 A0A1B6IKN7 A0A0C9R0A8 A0A1B6I2D9 E9J027 A0A1Y1JZA6 A0A0K8VY73 A0A1Y1JW70 A0A151J4G0 A0A026X3E6 A0A1Y1K0C5 A0A1Y1K3Y2 A0A0C9QDB0 A0A1Y1JW74 A0A1Y1JWS0 A0A3L8DPP1 A0A0C9RQN6 A0A0M8ZVL6 A0A1Y1JZA2 A0A1Y1K2G5 E0V9Q2 A0A1Y1K1M9 A0A1B6DB93 A0A1B6KAZ4 A0A1B6D5I0 J9K538 A0A1B6LLY0 A0A1B6MTV4 A0A1B6MHL2 A0A1B6CYQ6 A0A1B6LRU5 A0A1B6MPU4 A0A1B6DIY5 A0A1B6MSS5 A0A2P8Z2B5 A0A1B6L2X8 A0A0A1XK71 A0A0A1XNL3 B4K8S8 A0A2J7RE55 A0A0Q9X130 A0A154PBZ4 A0A0Q9X090 W8B5R1 A0A2J7RE59 A0A0Q9X293 A0A1A9WMZ4 A0A2J7RE60 A0A0Q9X0D1 V5JDH3 A0A0Q9XCJ2 A0A2M4CT47 A0A0K8VXG1 A0A0K8UZM7 A0A034VDU7 A0A0Q9X8U1 A0A034VIW0 A0A0K8VAP3 A0A0Q9XB37 W8AK98 A0A0Q9X0D3 A0A034VI82 A0A034VI86 A0A182W449 A0A1B0FAL3 A0A182Y544 A0A1A9VVG9 A0A034VHA6 A0A034VDV1 A0A0Q9X9U8 A0A034VG71 A0A3B0JCL4 A0A182PPX3 A0A182RH64 A0A2M4AMW3 A0A0Q9XCW9 F5HJ17 A0A034VIW6 A0A1E1W4F9
A0A139WKS9 A0A195EWW9 A0A158NB57 A0A139WL15 E2BYX3 A0A195D6L2 F4WXV5 A0A195BSR3 A0A151X0G5 A0A0L7QTH0 E2ARL7 A0A3S2LWV0 A0A2A3EMU1 A0A1B6HBC5 V5GSJ1 A0A088A0V7 A0A1B6IKN7 A0A0C9R0A8 A0A1B6I2D9 E9J027 A0A1Y1JZA6 A0A0K8VY73 A0A1Y1JW70 A0A151J4G0 A0A026X3E6 A0A1Y1K0C5 A0A1Y1K3Y2 A0A0C9QDB0 A0A1Y1JW74 A0A1Y1JWS0 A0A3L8DPP1 A0A0C9RQN6 A0A0M8ZVL6 A0A1Y1JZA2 A0A1Y1K2G5 E0V9Q2 A0A1Y1K1M9 A0A1B6DB93 A0A1B6KAZ4 A0A1B6D5I0 J9K538 A0A1B6LLY0 A0A1B6MTV4 A0A1B6MHL2 A0A1B6CYQ6 A0A1B6LRU5 A0A1B6MPU4 A0A1B6DIY5 A0A1B6MSS5 A0A2P8Z2B5 A0A1B6L2X8 A0A0A1XK71 A0A0A1XNL3 B4K8S8 A0A2J7RE55 A0A0Q9X130 A0A154PBZ4 A0A0Q9X090 W8B5R1 A0A2J7RE59 A0A0Q9X293 A0A1A9WMZ4 A0A2J7RE60 A0A0Q9X0D1 V5JDH3 A0A0Q9XCJ2 A0A2M4CT47 A0A0K8VXG1 A0A0K8UZM7 A0A034VDU7 A0A0Q9X8U1 A0A034VIW0 A0A0K8VAP3 A0A0Q9XB37 W8AK98 A0A0Q9X0D3 A0A034VI82 A0A034VI86 A0A182W449 A0A1B0FAL3 A0A182Y544 A0A1A9VVG9 A0A034VHA6 A0A034VDV1 A0A0Q9X9U8 A0A034VG71 A0A3B0JCL4 A0A182PPX3 A0A182RH64 A0A2M4AMW3 A0A0Q9XCW9 F5HJ17 A0A034VIW6 A0A1E1W4F9
Pubmed
EMBL
ODYU01002805
SOQ40756.1
KQ459220
KPJ02933.1
AGBW02012663
OWR44629.1
+ More
NNAY01000374 OXU28810.1 LJIG01002591 KRT84349.1 GGMS01001603 MBY70806.1 KQ971326 KYB28516.1 KQ981953 KYN32379.1 ADTU01010923 ADTU01010924 ADTU01010925 ADTU01010926 ADTU01010927 KYB28515.1 GL451531 EFN79091.1 KQ976760 KYN08513.1 GL888433 EGI60965.1 KQ976417 KYM89708.1 KQ982622 KYQ53714.1 KQ414744 KOC61920.1 GL442153 EFN63925.1 RSAL01000148 RVE45822.1 KZ288208 PBC33027.1 GECU01035756 JAS71950.1 GALX01003894 JAB64572.1 GECU01020214 JAS87492.1 GBYB01009549 JAG79316.1 GECU01026610 GECU01019727 JAS81096.1 JAS87979.1 GL767291 EFZ13795.1 GEZM01099014 JAV53571.1 GDHF01033681 GDHF01008502 JAI18633.1 JAI43812.1 GEZM01099023 JAV53564.1 KQ980151 KYN17522.1 KK107039 EZA61919.1 GEZM01099017 JAV53570.1 GEZM01099020 JAV53567.1 GBYB01001269 JAG71036.1 GEZM01099018 JAV53569.1 GEZM01099024 JAV53563.1 QOIP01000006 RLU21788.1 GBYB01009551 JAG79318.1 KQ435859 KOX70449.1 GEZM01099019 JAV53568.1 GEZM01099021 JAV53566.1 DS234995 EEB10087.1 GEZM01099022 JAV53565.1 GEDC01014360 JAS22938.1 GEBQ01031359 GEBQ01022724 JAT08618.1 JAT17253.1 GEDC01016360 JAS20938.1 ABLF02031390 GEBQ01015319 JAT24658.1 GEBQ01029673 GEBQ01029473 GEBQ01024353 GEBQ01019330 GEBQ01002485 GEBQ01000652 JAT10304.1 JAT10504.1 JAT15624.1 JAT20647.1 JAT37492.1 JAT39325.1 GEBQ01004571 JAT35406.1 GEDC01018843 GEDC01001079 JAS18455.1 JAS36219.1 GEBQ01013630 JAT26347.1 GEBQ01025905 GEBQ01006430 GEBQ01002092 JAT14072.1 JAT33547.1 JAT37885.1 GEDC01025552 GEDC01019828 GEDC01011637 JAS11746.1 JAS17470.1 JAS25661.1 GEBQ01001011 JAT38966.1 PYGN01000229 PSN50637.1 GEBQ01023878 GEBQ01021918 JAT16099.1 JAT18059.1 GBXI01002528 JAD11764.1 GBXI01001827 JAD12465.1 CH933806 EDW15497.1 NEVH01005277 PNF39116.1 KRG01589.1 KQ434869 KZC09439.1 KRG01583.1 GAMC01021416 JAB85139.1 PNF39119.1 KRG01593.1 PNF39117.1 KRG01584.1 KC122909 AGI96987.1 KRG01592.1 GGFL01004348 MBW68526.1 GDHF01008737 JAI43577.1 GDHF01020187 JAI32127.1 GAKP01017456 JAC41496.1 KRG01588.1 GAKP01017454 JAC41498.1 GDHF01016412 JAI35902.1 KRG01591.1 GAMC01021421 GAMC01021419 GAMC01021417 JAB85138.1 KRG01590.1 GAKP01017452 JAC41500.1 GAKP01017447 JAC41505.1 CCAG010006880 GAKP01017450 JAC41502.1 GAKP01017451 JAC41501.1 KRG01586.1 GAKP01017459 GAKP01017448 JAC41504.1 OUUW01000005 SPP80114.1 GGFK01008800 MBW42121.1 KRG01587.1 AAAB01008811 EGK96281.1 GAKP01017455 GAKP01017449 JAC41503.1 GDQN01009188 JAT81866.1
NNAY01000374 OXU28810.1 LJIG01002591 KRT84349.1 GGMS01001603 MBY70806.1 KQ971326 KYB28516.1 KQ981953 KYN32379.1 ADTU01010923 ADTU01010924 ADTU01010925 ADTU01010926 ADTU01010927 KYB28515.1 GL451531 EFN79091.1 KQ976760 KYN08513.1 GL888433 EGI60965.1 KQ976417 KYM89708.1 KQ982622 KYQ53714.1 KQ414744 KOC61920.1 GL442153 EFN63925.1 RSAL01000148 RVE45822.1 KZ288208 PBC33027.1 GECU01035756 JAS71950.1 GALX01003894 JAB64572.1 GECU01020214 JAS87492.1 GBYB01009549 JAG79316.1 GECU01026610 GECU01019727 JAS81096.1 JAS87979.1 GL767291 EFZ13795.1 GEZM01099014 JAV53571.1 GDHF01033681 GDHF01008502 JAI18633.1 JAI43812.1 GEZM01099023 JAV53564.1 KQ980151 KYN17522.1 KK107039 EZA61919.1 GEZM01099017 JAV53570.1 GEZM01099020 JAV53567.1 GBYB01001269 JAG71036.1 GEZM01099018 JAV53569.1 GEZM01099024 JAV53563.1 QOIP01000006 RLU21788.1 GBYB01009551 JAG79318.1 KQ435859 KOX70449.1 GEZM01099019 JAV53568.1 GEZM01099021 JAV53566.1 DS234995 EEB10087.1 GEZM01099022 JAV53565.1 GEDC01014360 JAS22938.1 GEBQ01031359 GEBQ01022724 JAT08618.1 JAT17253.1 GEDC01016360 JAS20938.1 ABLF02031390 GEBQ01015319 JAT24658.1 GEBQ01029673 GEBQ01029473 GEBQ01024353 GEBQ01019330 GEBQ01002485 GEBQ01000652 JAT10304.1 JAT10504.1 JAT15624.1 JAT20647.1 JAT37492.1 JAT39325.1 GEBQ01004571 JAT35406.1 GEDC01018843 GEDC01001079 JAS18455.1 JAS36219.1 GEBQ01013630 JAT26347.1 GEBQ01025905 GEBQ01006430 GEBQ01002092 JAT14072.1 JAT33547.1 JAT37885.1 GEDC01025552 GEDC01019828 GEDC01011637 JAS11746.1 JAS17470.1 JAS25661.1 GEBQ01001011 JAT38966.1 PYGN01000229 PSN50637.1 GEBQ01023878 GEBQ01021918 JAT16099.1 JAT18059.1 GBXI01002528 JAD11764.1 GBXI01001827 JAD12465.1 CH933806 EDW15497.1 NEVH01005277 PNF39116.1 KRG01589.1 KQ434869 KZC09439.1 KRG01583.1 GAMC01021416 JAB85139.1 PNF39119.1 KRG01593.1 PNF39117.1 KRG01584.1 KC122909 AGI96987.1 KRG01592.1 GGFL01004348 MBW68526.1 GDHF01008737 JAI43577.1 GDHF01020187 JAI32127.1 GAKP01017456 JAC41496.1 KRG01588.1 GAKP01017454 JAC41498.1 GDHF01016412 JAI35902.1 KRG01591.1 GAMC01021421 GAMC01021419 GAMC01021417 JAB85138.1 KRG01590.1 GAKP01017452 JAC41500.1 GAKP01017447 JAC41505.1 CCAG010006880 GAKP01017450 JAC41502.1 GAKP01017451 JAC41501.1 KRG01586.1 GAKP01017459 GAKP01017448 JAC41504.1 OUUW01000005 SPP80114.1 GGFK01008800 MBW42121.1 KRG01587.1 AAAB01008811 EGK96281.1 GAKP01017455 GAKP01017449 JAC41503.1 GDQN01009188 JAT81866.1
Proteomes
UP000053268
UP000007151
UP000215335
UP000007266
UP000078541
UP000005205
+ More
UP000008237 UP000078542 UP000007755 UP000078540 UP000075809 UP000053825 UP000000311 UP000283053 UP000242457 UP000005203 UP000078492 UP000053097 UP000279307 UP000053105 UP000009046 UP000007819 UP000245037 UP000009192 UP000235965 UP000076502 UP000091820 UP000075920 UP000092444 UP000076408 UP000078200 UP000268350 UP000075885 UP000075900 UP000007062
UP000008237 UP000078542 UP000007755 UP000078540 UP000075809 UP000053825 UP000000311 UP000283053 UP000242457 UP000005203 UP000078492 UP000053097 UP000279307 UP000053105 UP000009046 UP000007819 UP000245037 UP000009192 UP000235965 UP000076502 UP000091820 UP000075920 UP000092444 UP000076408 UP000078200 UP000268350 UP000075885 UP000075900 UP000007062
Pfam
PF03250 Tropomodulin
Gene 3D
ProteinModelPortal
A0A2H1VIY4
A0A194QBL0
A0A212ET25
A0A232FE74
A0A0T6BB80
A0A2S2PZ53
+ More
A0A139WKS9 A0A195EWW9 A0A158NB57 A0A139WL15 E2BYX3 A0A195D6L2 F4WXV5 A0A195BSR3 A0A151X0G5 A0A0L7QTH0 E2ARL7 A0A3S2LWV0 A0A2A3EMU1 A0A1B6HBC5 V5GSJ1 A0A088A0V7 A0A1B6IKN7 A0A0C9R0A8 A0A1B6I2D9 E9J027 A0A1Y1JZA6 A0A0K8VY73 A0A1Y1JW70 A0A151J4G0 A0A026X3E6 A0A1Y1K0C5 A0A1Y1K3Y2 A0A0C9QDB0 A0A1Y1JW74 A0A1Y1JWS0 A0A3L8DPP1 A0A0C9RQN6 A0A0M8ZVL6 A0A1Y1JZA2 A0A1Y1K2G5 E0V9Q2 A0A1Y1K1M9 A0A1B6DB93 A0A1B6KAZ4 A0A1B6D5I0 J9K538 A0A1B6LLY0 A0A1B6MTV4 A0A1B6MHL2 A0A1B6CYQ6 A0A1B6LRU5 A0A1B6MPU4 A0A1B6DIY5 A0A1B6MSS5 A0A2P8Z2B5 A0A1B6L2X8 A0A0A1XK71 A0A0A1XNL3 B4K8S8 A0A2J7RE55 A0A0Q9X130 A0A154PBZ4 A0A0Q9X090 W8B5R1 A0A2J7RE59 A0A0Q9X293 A0A1A9WMZ4 A0A2J7RE60 A0A0Q9X0D1 V5JDH3 A0A0Q9XCJ2 A0A2M4CT47 A0A0K8VXG1 A0A0K8UZM7 A0A034VDU7 A0A0Q9X8U1 A0A034VIW0 A0A0K8VAP3 A0A0Q9XB37 W8AK98 A0A0Q9X0D3 A0A034VI82 A0A034VI86 A0A182W449 A0A1B0FAL3 A0A182Y544 A0A1A9VVG9 A0A034VHA6 A0A034VDV1 A0A0Q9X9U8 A0A034VG71 A0A3B0JCL4 A0A182PPX3 A0A182RH64 A0A2M4AMW3 A0A0Q9XCW9 F5HJ17 A0A034VIW6 A0A1E1W4F9
A0A139WKS9 A0A195EWW9 A0A158NB57 A0A139WL15 E2BYX3 A0A195D6L2 F4WXV5 A0A195BSR3 A0A151X0G5 A0A0L7QTH0 E2ARL7 A0A3S2LWV0 A0A2A3EMU1 A0A1B6HBC5 V5GSJ1 A0A088A0V7 A0A1B6IKN7 A0A0C9R0A8 A0A1B6I2D9 E9J027 A0A1Y1JZA6 A0A0K8VY73 A0A1Y1JW70 A0A151J4G0 A0A026X3E6 A0A1Y1K0C5 A0A1Y1K3Y2 A0A0C9QDB0 A0A1Y1JW74 A0A1Y1JWS0 A0A3L8DPP1 A0A0C9RQN6 A0A0M8ZVL6 A0A1Y1JZA2 A0A1Y1K2G5 E0V9Q2 A0A1Y1K1M9 A0A1B6DB93 A0A1B6KAZ4 A0A1B6D5I0 J9K538 A0A1B6LLY0 A0A1B6MTV4 A0A1B6MHL2 A0A1B6CYQ6 A0A1B6LRU5 A0A1B6MPU4 A0A1B6DIY5 A0A1B6MSS5 A0A2P8Z2B5 A0A1B6L2X8 A0A0A1XK71 A0A0A1XNL3 B4K8S8 A0A2J7RE55 A0A0Q9X130 A0A154PBZ4 A0A0Q9X090 W8B5R1 A0A2J7RE59 A0A0Q9X293 A0A1A9WMZ4 A0A2J7RE60 A0A0Q9X0D1 V5JDH3 A0A0Q9XCJ2 A0A2M4CT47 A0A0K8VXG1 A0A0K8UZM7 A0A034VDU7 A0A0Q9X8U1 A0A034VIW0 A0A0K8VAP3 A0A0Q9XB37 W8AK98 A0A0Q9X0D3 A0A034VI82 A0A034VI86 A0A182W449 A0A1B0FAL3 A0A182Y544 A0A1A9VVG9 A0A034VHA6 A0A034VDV1 A0A0Q9X9U8 A0A034VG71 A0A3B0JCL4 A0A182PPX3 A0A182RH64 A0A2M4AMW3 A0A0Q9XCW9 F5HJ17 A0A034VIW6 A0A1E1W4F9
PDB
1PGV
E-value=3.39921e-33,
Score=351
Ontologies
PANTHER
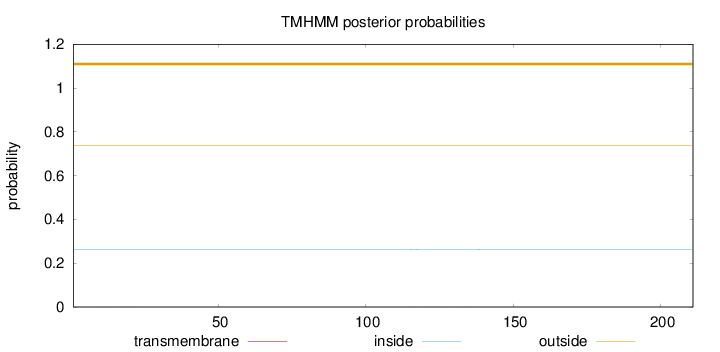
Topology
Length:
211
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01545
Exp number, first 60 AAs:
0.00253
Total prob of N-in:
0.26261
outside
1 - 211
Population Genetic Test Statistics
Pi
246.850629
Theta
177.798338
Tajima's D
2.031881
CLR
0.548344
CSRT
0.887355632218389
Interpretation
Uncertain
