Gene
KWMTBOMO05295 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002508
Annotation
PREDICTED:_pyruvate_kinase-like_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.367
Sequence
CDS
ATGCCATTTAATTTTGTAACCGAAACGAAAAAAGGTGTCGATAAACCAAGATTCTCACTATCACTGAACAGAAATAATGCGCCACATTTGAGCAACGAAAACAAGAAAAATAACGAGAATATTAAAGAGAATACAGATCCATGCAGTTCTGGTAATAACAAGGAGAATAAGCCATTAAAGAGGCCGCTCAAAAGCATATATTCCAACACGTTGAGTGCAGCAAAAAAATTGAAGCCTCTAAGTGAATCATCGAAATCTGCAGGTCCCACAGTAAATAGGGACACAGCAACCCTAAAGCATTCAGACAGAACATTGAATCCAGAGCTAGCAATTCTGGAGGAGGGAGAATCAAGCTCGGTCATGTTGAATGGCCACCACCCTCCTGACACTCAATATGGAGGGGTGCAGAAGAAGATGCCTGTGTCTTCACTGTCCAATTTGGGGAACACTTGTTTTCTCAATAGTGTGCTTTATACTTTGAGGTATGCACCTCGGTTCCTGCACAACCTTCACCATTTAGTGTCAGATCTGGCCAGTGTTGAGCAAAAGTTGGGCAGCATCAGGCTCAAAAGTTCTTCATTGGGTCGGAGTGCTGCGGGCTTAGTATCATCTGGGACTAGATCATGGAGTAGCAAGGACTTGTTGTCATTAGGCCAGTCAGATAATAGTTCTGGAAAAAGTAAAATACAGATAGCGACAGAAAAACTTCACGAGACCTACTTTAATCTTCGTGCAGCTGAGAACAAATGTCTGAACAGCACCAATTCTGAGGCTACCCCTGAGCCATATGCAGCGGATGCCTTCTTGGCTGCTCTTAGAGATGTTAATTCTACTTTTGAAGGTAATCGACAGCAGGACGCTCATGAATTATTAGTTTGTATTTTGGACAACATACGGGAGACCTGCAGAGCGCTGAGTGCGCGCGCTGCTCGACTACAGATGCACGAAAACGGAGACAGTAACGGCATAGGCCGCCAGCCAAGCTTAGATGGAGACAACGGGAAACCAACATTAGGAAATCTTCGTAAGTCGTGGAAGAAACGCAAAGAAATCAAGGCAATCAACGACAAAAGGAATTCTCCGACTGAAGAACGGGAACCGTCGCCTGTGGATGCTGAGAAAGATGAGCGGTCGAGACCAGGATGGGACTTTGTGGCTGAAGATTTTGAAGGTACGATGGTAGTGCGGACCATGTGCTTAGAATGTGAAGCGGTCACTCAGAAGGCGCAGGCTGTTTGTGAGTTGTGCGTTCCGGTTGGCGAAGAAGAACCAGGAGCGGAACCGTTCCGGGCGGCCTGCCTGTCTACAGAATACCTGCGTGATCAGAACAAGTATTGGTGCGAACGTTGTCTACGGTACAACGAAGCTCGTCGCAGCGTCACTTATTCGAGACTGCCGCGGTTGCTGGTACTACAGTTGAAACGATTCAGCGGGGGAATGGAAAAGATAACAAGACATGCACCGACGCCTCTACTTATGCCTTGTTTCTGTGAGCCTTGCGCTTCCAGACCGCCCGAGCGTGCACCACAGCACAGATACATCCTATGGGCAGTGATAATGCACCTCGGTCAGACCCTAACAGGCGGGCATTACGTTGCTTATGCGCGCGATAGTTCTTGTGACTTCAAGTGCTCTCGTGAAGGTAGCGGACGGTTCGCGCTCTCCCGACGTGGAATTCAATTTTTCACTCGGAAGGGCAGTTCAAGATCACGTAAACCGCCCGTCAGTCGTAACCGAGAGCTCACAGACAAATATCTTCACTATAATCCATTAAAAATGGTTTACCCTACTATTCACGATGGCGAAATCGAGGCGGGTGCAGTTGAAAAGCCCACAGTCGCCAACGTCGGCTCCCAGCTCCAGCATATGTGCGGTCTCGACATTGACTCCAAGTCTTCGTACATCCGTCTCTCCGGAATAATCTGTACTATTGGACCGGCATCCCGTAATGTAGCTGTGCTAGAGAAGATGATGGAGACGGGAATGAACGTGGCAAGGATGAACTTCTCGCACGGTTCCCATGAGTACCACGCAGAGACCATTCGCAATTGTCGCGAGGCCGAGAAGAGTTACAGCGCTAAACTAGGTTCACCCTTCTCTTTGGCCATCGCATTGGACACTAAAGGACCTGAGATCAGAACTGGACTCTTGGAAGGCGGCGGCTCAGCTGAAGTCGAACTGAAGAAGGGAGAGACTATAAAGTTGACGACAAGTTCGGATTATCAAGAGAAAGGCAATGCTGATACAATTTACGTGGACTACAAGAACATAACGAATGTCGTGAAGCCAGGAAACCGAATCTTCATCGATGATGGCCTCATCTCTATCATCTGTCAGTCGGTCAGCGCTGACACTCTTACGTGTACCATTGAAAACGGAGGTATGCTCGGATCCCGGAAAGGCGTCAACCTGCCCGGCATACCCGTGGACCTACCCGCAGTCTCCGAGAAGGACAAGTCGGACCTGCTCTTCGGCGTCGAACAAGGCGTTGATATGATTTTCGCGTCATTCATCCGCAACGGCGCCGCGCTGCACGAGATTCGCGGCATCCTCGGCGAAAAAGGCAAGAACATCAAGATCATCTCCAAGATCGAAAATCACCAGGGAATGGTCAATTTAGACGAGATTATAGCTGAATCCGATGGTATCATGGTTGCTCGCGGAGATCTGGGTATCGAGATCCCTCCAGAAAAGGTATTCCTCGCCCAGAAGACCATGATCGCCAGATGCAATCGGGTTGGAAAACCAGTGATCTGTGCGACTCAAATGCTGGAATCCATGGTTAAGAAGCCCCGTCCCACCAGAGCCGAGATCTCTGACGTGGCCAACGCCATCCTCGACGGAGCTGACTGCGTGATGCTCTCCGGGGAGACTGCCAAGGGCGACTATCCTGTCGAGTGTGTTCACACCATGGCCAACATTTGCAAGGAGGCTGAAGCTGTAATCTGGCACAGGCAACTCTTCAATGACCTTGTGTCCGAGGTGAAGCCGCCGATCGATCCCGCGCACTCGGCCGCCATCGCCGCGGTGGAGGCGGCCACCAAGTGCCTGGCGTCGGCCATCGTGGTCATCACGACGAGCGGCAAGTCGGCGCACCTGCTCAGCAAGTACCGGCCGCGCTGCCCCATCATCGCCGTCACCAGGCACCCGCAGACCGCCAGGCAGGTGCACCTCTACCGCGGCGTGCTGCCCATCGTCTACCAAGAGCCGACGGCGTCGGACTGGCTGAAGGACGTGGACAACCGCGTGCAGAGCGGGCTGCGGTTCGGGCGGCAGCGCGGCTTCGTGCACCCCGGGGACAACGCGGTCGTCGTCACCGGCTGGAAGCAGGGCTCCGGCTTCACCAACACCGTCCGGGTCATTCAATTGGAATAA
Protein
MPFNFVTETKKGVDKPRFSLSLNRNNAPHLSNENKKNNENIKENTDPCSSGNNKENKPLKRPLKSIYSNTLSAAKKLKPLSESSKSAGPTVNRDTATLKHSDRTLNPELAILEEGESSSVMLNGHHPPDTQYGGVQKKMPVSSLSNLGNTCFLNSVLYTLRYAPRFLHNLHHLVSDLASVEQKLGSIRLKSSSLGRSAAGLVSSGTRSWSSKDLLSLGQSDNSSGKSKIQIATEKLHETYFNLRAAENKCLNSTNSEATPEPYAADAFLAALRDVNSTFEGNRQQDAHELLVCILDNIRETCRALSARAARLQMHENGDSNGIGRQPSLDGDNGKPTLGNLRKSWKKRKEIKAINDKRNSPTEEREPSPVDAEKDERSRPGWDFVAEDFEGTMVVRTMCLECEAVTQKAQAVCELCVPVGEEEPGAEPFRAACLSTEYLRDQNKYWCERCLRYNEARRSVTYSRLPRLLVLQLKRFSGGMEKITRHAPTPLLMPCFCEPCASRPPERAPQHRYILWAVIMHLGQTLTGGHYVAYARDSSCDFKCSREGSGRFALSRRGIQFFTRKGSSRSRKPPVSRNRELTDKYLHYNPLKMVYPTIHDGEIEAGAVEKPTVANVGSQLQHMCGLDIDSKSSYIRLSGIICTIGPASRNVAVLEKMMETGMNVARMNFSHGSHEYHAETIRNCREAEKSYSAKLGSPFSLAIALDTKGPEIRTGLLEGGGSAEVELKKGETIKLTTSSDYQEKGNADTIYVDYKNITNVVKPGNRIFIDDGLISIICQSVSADTLTCTIENGGMLGSRKGVNLPGIPVDLPAVSEKDKSDLLFGVEQGVDMIFASFIRNGAALHEIRGILGEKGKNIKIISKIENHQGMVNLDEIIAESDGIMVARGDLGIEIPPEKVFLAQKTMIARCNRVGKPVICATQMLESMVKKPRPTRAEISDVANAILDGADCVMLSGETAKGDYPVECVHTMANICKEAEAVIWHRQLFNDLVSEVKPPIDPAHSAAIAAVEAATKCLASAIVVITTSGKSAHLLSKYRPRCPIIAVTRHPQTARQVHLYRGVLPIVYQEPTASDWLKDVDNRVQSGLRFGRQRGFVHPGDNAVVVTGWKQGSGFTNTVRVIQLE
Summary
Uniprot
ProteinModelPortal
PDB
6DU6
E-value=0,
Score=2038
Ontologies
GO
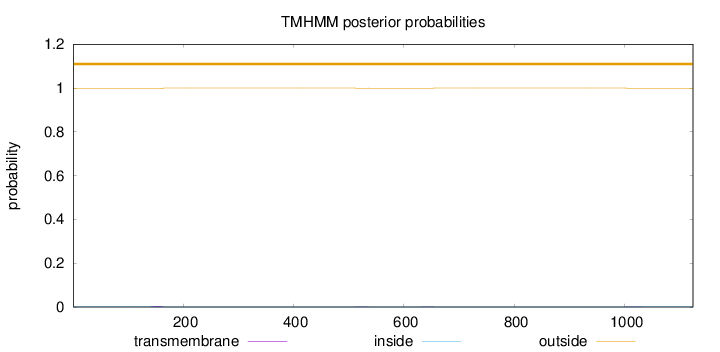
Topology
Length:
1124
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1092
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00306
outside
1 - 1124
Population Genetic Test Statistics
Pi
229.138981
Theta
178.401601
Tajima's D
1.058075
CLR
0.11888
CSRT
0.672816359182041
Interpretation
Uncertain
