Gene
KWMTBOMO05287
Pre Gene Modal
BGIBMGA002402
Annotation
PREDICTED:_sorting_nexin-4-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.356 Nuclear Reliability : 1.744
Sequence
CDS
ATGTCTAAAGATAATACACCGGTCAAATCACCTAAATCGCCAAAAGCAATAAAAACAGTAGAAGACTTCGAAAATCAGGACACACTGTTGAAGCATGTCGATATAACTATTGTGGAGTCGGAGAAACGAGCTAATGGTACTCTACACGTCAGAGATCATTACACAGTATATCTCGTCGATTTAAAAGTTACAGATCCTGATTACAAATTAGTACAGTCAAAAATAAGCACAATCTGGAGGAGATACACTGAATTCGAACAAATACATGATTACCTACAAGTCACATATCCACATGTTGTTATACCTCCATTACCAGAGAAAAGGGTGCTTTACGCGTGGCGCAAATCGGACACTACAGACCCGGAGTTTGTGGAGCGCCGTCGAGCTGGCTTAGAAAATTTCCTATTACGAGTCGCATCTCACCCTCGACTATGTTTCGATGATCAATTCATTAATTTCCTACAAGAAGAACATGGTTGGCGGGAAACTATCACCGATAGTGGATATTTACTACAAGCTGAAAATAAAATAAAATCACTGTCCGTATCTATAAGGTTGAAGAAGCCCGATCCTGAGGTTGAGTCCTTCAAAAACTACGGGAAACAATTAGAAACTAATTTAGGAAACTTTTTATATACAAGATCCAAAATAATAGAAAAAAATTACGCTCTATGTAAACTGCACGCGAATTACGGCAAACTGTTTAGTGAATGGAGCGTCATCGAGAAGGAGATGGGAGACGGATTGCAGAAGGCCGGACATTATTTCGATTCAATAGCGGACAGTATAGATTCGGTTGCGGAAGACGAAGAACAGCTGGCGGACCAATTAAAAGAGTATCTGTTTTATGCGGGAGCTCTGCAGCAGCTGTGCAGCAATCATGAGGCTCTCCAACGAAACCTGGAACAGGCACAAGACACGCTCAATAATCGGTTAGCAGAACGCACCAGAGCGGCGGCGGGAAAATCCGGTATAATGTCCCGACTGTTTGGTACCACGGATCCTGATATAGTGCGGGACCAAACTACGCGAGCTCTCGACGCTAAGATACAAGCAGATAACGAAGCTATAGAAAAGGCTAAACTGGATTTGGAAGAATTCACAAAGAAAGTTATGGTAGAAATAGACCATTTCCAGAAGCAAAAAGATAAGGACTTGCACGAGTCGCTAGTAAATTTCATAACTTTACAAGTTAAAGCTGCGAAAAGGAATTTGCAAGCATGGACACAAATAAAAGAATGTCTACAGAATATGCCTTAA
Protein
MSKDNTPVKSPKSPKAIKTVEDFENQDTLLKHVDITIVESEKRANGTLHVRDHYTVYLVDLKVTDPDYKLVQSKISTIWRRYTEFEQIHDYLQVTYPHVVIPPLPEKRVLYAWRKSDTTDPEFVERRRAGLENFLLRVASHPRLCFDDQFINFLQEEHGWRETITDSGYLLQAENKIKSLSVSIRLKKPDPEVESFKNYGKQLETNLGNFLYTRSKIIEKNYALCKLHANYGKLFSEWSVIEKEMGDGLQKAGHYFDSIADSIDSVAEDEEQLADQLKEYLFYAGALQQLCSNHEALQRNLEQAQDTLNNRLAERTRAAAGKSGIMSRLFGTTDPDIVRDQTTRALDAKIQADNEAIEKAKLDLEEFTKKVMVEIDHFQKQKDKDLHESLVNFITLQVKAAKRNLQAWTQIKECLQNMP
Summary
Uniprot
H9IYR8
A0A2H1WMU9
A0A2A4JRU7
A0A212ET07
A0A0N0PEA9
A0A194QBK0
+ More
A0A2J7RK00 V5G7V2 K7ITX5 A0A232F0I9 E2ADZ0 D6W9D7 A0A154PJI0 A0A088A3A8 A0A026W4X0 A0A2A3E7H9 A0A0T6B6D3 E2BCE6 A0A0C9QGZ7 A0A310SL48 A0A158NBE2 A0A1W4WCU7 A0A0N0BCJ6 A0A1Y1KL99 U4U7A6 N6T9R4 A0A1B6J6D7 A0A195BAD5 E9IRA8 F4X8U7 A0A195FCQ1 A0A151IL05 E0VR31 A0A2P8ZGZ8 A0A195DRN1 A0A0C9RHA9 A0A1B6KH74 A0A0V0G930 A0A069DTZ1 A0A023F0T5 A0A1U7R678 A0A1U7RDE9 A0A1S3IHE9 A0A0P5K093 A0A0P5FVX3 A0A3B4DSY9 A0A2I0MG79 W5ULJ2 A0A0A9W7E4 A0A091MBQ8 A0A1V4K5Z3 A0A091I5C9 B0S5M0 R0LM96 W5KF39 K7F6Y1 A0A099YX94 Q58EC1 A0A091VHJ0 A0A093PAK9 A0A087QNP4 A0A091M0W4 A0A091GY55 A0A091UL85 A0A094K8J8 A0A093JVH6 A0A0A9YCE9 A0A087VPU8 A0A091QG71 A0A091JD49 U3KGG0 H0Z0N2 A0A2T7PR86 A0A093IUE0 A0A093CA41 A0A093IMF1 A0A1A7XU72 A0A3Q2U656 A0A3B4GUN0 A0A3Q4HLC0 A0A3P8Q918 A0A3Q2G300 A0A1A7YFH9 A0A091QAI3 G1NHV5 E1BQF6 A0A0F7ZCN0 A0A091UHJ8 A0A091RKD6 A0A091RSY9 A0A091F098 A0A3Q2UUK5 A0A3B3T8K6 A0A0P7U4Q6 U3INH1 I3J8N9 G1T9G4 A0A093QN06 K1QVP1 A0A060W9R2
A0A2J7RK00 V5G7V2 K7ITX5 A0A232F0I9 E2ADZ0 D6W9D7 A0A154PJI0 A0A088A3A8 A0A026W4X0 A0A2A3E7H9 A0A0T6B6D3 E2BCE6 A0A0C9QGZ7 A0A310SL48 A0A158NBE2 A0A1W4WCU7 A0A0N0BCJ6 A0A1Y1KL99 U4U7A6 N6T9R4 A0A1B6J6D7 A0A195BAD5 E9IRA8 F4X8U7 A0A195FCQ1 A0A151IL05 E0VR31 A0A2P8ZGZ8 A0A195DRN1 A0A0C9RHA9 A0A1B6KH74 A0A0V0G930 A0A069DTZ1 A0A023F0T5 A0A1U7R678 A0A1U7RDE9 A0A1S3IHE9 A0A0P5K093 A0A0P5FVX3 A0A3B4DSY9 A0A2I0MG79 W5ULJ2 A0A0A9W7E4 A0A091MBQ8 A0A1V4K5Z3 A0A091I5C9 B0S5M0 R0LM96 W5KF39 K7F6Y1 A0A099YX94 Q58EC1 A0A091VHJ0 A0A093PAK9 A0A087QNP4 A0A091M0W4 A0A091GY55 A0A091UL85 A0A094K8J8 A0A093JVH6 A0A0A9YCE9 A0A087VPU8 A0A091QG71 A0A091JD49 U3KGG0 H0Z0N2 A0A2T7PR86 A0A093IUE0 A0A093CA41 A0A093IMF1 A0A1A7XU72 A0A3Q2U656 A0A3B4GUN0 A0A3Q4HLC0 A0A3P8Q918 A0A3Q2G300 A0A1A7YFH9 A0A091QAI3 G1NHV5 E1BQF6 A0A0F7ZCN0 A0A091UHJ8 A0A091RKD6 A0A091RSY9 A0A091F098 A0A3Q2UUK5 A0A3B3T8K6 A0A0P7U4Q6 U3INH1 I3J8N9 G1T9G4 A0A093QN06 K1QVP1 A0A060W9R2
Pubmed
19121390
22118469
26354079
20075255
28648823
20798317
+ More
18362917 19820115 24508170 30249741 21347285 28004739 23537049 21282665 21719571 20566863 29403074 26334808 25474469 23371554 23127152 25401762 23594743 25329095 17381049 20360741 15592404 25186727 20838655 29240929 23749191 21993624 22992520 24755649
18362917 19820115 24508170 30249741 21347285 28004739 23537049 21282665 21719571 20566863 29403074 26334808 25474469 23371554 23127152 25401762 23594743 25329095 17381049 20360741 15592404 25186727 20838655 29240929 23749191 21993624 22992520 24755649
EMBL
BABH01010921
BABH01010922
ODYU01009391
SOQ53784.1
NWSH01000697
PCG74731.1
+ More
AGBW02012663 OWR44635.1 KQ459927 KPJ19312.1 KQ459220 KPJ02923.1 NEVH01002981 PNF41162.1 GALX01002266 JAB66200.1 NNAY01001414 OXU24053.1 GL438827 EFN68223.1 KQ971312 EEZ98174.1 KQ434938 KZC12041.1 KK107453 QOIP01000004 EZA50651.1 RLU23873.1 KZ288354 PBC27242.1 LJIG01009514 KRT82928.1 GL447257 EFN86682.1 GBYB01013903 JAG83670.1 KQ760133 OAD61787.1 ADTU01010992 ADTU01010993 KQ435900 KOX69092.1 GEZM01080406 GEZM01080405 GEZM01080404 GEZM01080403 GEZM01080402 GEZM01080401 JAV62169.1 KB631815 ERL86491.1 APGK01046266 KB741050 ENN74478.1 GECU01012967 JAS94739.1 KQ976540 KYM81187.1 GL765107 EFZ16914.1 GL888932 EGI57366.1 KQ981673 KYN38171.1 KQ977149 KYN05297.1 DS235442 EEB15837.1 PYGN01000060 PSN55740.1 KQ980581 KYN15462.1 GBYB01012592 JAG82359.1 GEBQ01029177 JAT10800.1 GECL01001574 JAP04550.1 GBGD01001727 JAC87162.1 GBBI01004096 JAC14616.1 GDIP01151000 GDIQ01191666 LRGB01000007 JAJ72402.1 JAK60059.1 KZS21960.1 GDIQ01256770 JAJ94954.1 AKCR02000014 PKK28699.1 JT418917 AHH42892.1 GBHO01039910 GBRD01006640 JAG03694.1 JAG59181.1 KK823593 KFP71587.1 LSYS01004331 OPJ79801.1 KL218243 KFP03382.1 BX255930 KB742929 EOB02815.1 AGCU01193299 AGCU01193300 AGCU01193301 KL887308 KGL74829.1 BC091982 BC164735 AAH91982.1 AAI64735.1 KK733996 KFR02235.1 KL225231 KFW69282.1 KL225768 KFM02848.1 KK514504 KFP64282.1 KL513910 KFO87360.1 KL409758 KFQ91699.1 KL339221 KFZ51841.1 KL206458 KFV82544.1 GBHO01014303 JAG29301.1 KL501232 KFO14640.1 KK696997 KFQ24952.1 KK501769 KFP17635.1 AGTO01000332 ABQF01011049 PZQS01000002 PVD35887.1 KL216593 KFV70296.1 KL460806 KFV12820.1 KK590766 KFV99933.1 HADW01020168 SBP21568.1 AADN05000350 HADX01006746 SBP28978.1 KK690812 KFQ16566.1 GBEX01002226 JAI12334.1 KK435974 KFQ90244.1 KK820088 KFQ39737.1 KK932664 KFQ45770.1 KK719330 KFO62656.1 JARO02008923 KPP62163.1 ADON01058539 ADON01058540 AERX01013232 AAGW02013744 AAGW02013745 AAGW02013746 AAGW02013747 AAGW02013748 AAGW02013749 AAGW02013750 KL425962 KFW90283.1 JH816117 EKC33010.1 FR904394 CDQ61999.1
AGBW02012663 OWR44635.1 KQ459927 KPJ19312.1 KQ459220 KPJ02923.1 NEVH01002981 PNF41162.1 GALX01002266 JAB66200.1 NNAY01001414 OXU24053.1 GL438827 EFN68223.1 KQ971312 EEZ98174.1 KQ434938 KZC12041.1 KK107453 QOIP01000004 EZA50651.1 RLU23873.1 KZ288354 PBC27242.1 LJIG01009514 KRT82928.1 GL447257 EFN86682.1 GBYB01013903 JAG83670.1 KQ760133 OAD61787.1 ADTU01010992 ADTU01010993 KQ435900 KOX69092.1 GEZM01080406 GEZM01080405 GEZM01080404 GEZM01080403 GEZM01080402 GEZM01080401 JAV62169.1 KB631815 ERL86491.1 APGK01046266 KB741050 ENN74478.1 GECU01012967 JAS94739.1 KQ976540 KYM81187.1 GL765107 EFZ16914.1 GL888932 EGI57366.1 KQ981673 KYN38171.1 KQ977149 KYN05297.1 DS235442 EEB15837.1 PYGN01000060 PSN55740.1 KQ980581 KYN15462.1 GBYB01012592 JAG82359.1 GEBQ01029177 JAT10800.1 GECL01001574 JAP04550.1 GBGD01001727 JAC87162.1 GBBI01004096 JAC14616.1 GDIP01151000 GDIQ01191666 LRGB01000007 JAJ72402.1 JAK60059.1 KZS21960.1 GDIQ01256770 JAJ94954.1 AKCR02000014 PKK28699.1 JT418917 AHH42892.1 GBHO01039910 GBRD01006640 JAG03694.1 JAG59181.1 KK823593 KFP71587.1 LSYS01004331 OPJ79801.1 KL218243 KFP03382.1 BX255930 KB742929 EOB02815.1 AGCU01193299 AGCU01193300 AGCU01193301 KL887308 KGL74829.1 BC091982 BC164735 AAH91982.1 AAI64735.1 KK733996 KFR02235.1 KL225231 KFW69282.1 KL225768 KFM02848.1 KK514504 KFP64282.1 KL513910 KFO87360.1 KL409758 KFQ91699.1 KL339221 KFZ51841.1 KL206458 KFV82544.1 GBHO01014303 JAG29301.1 KL501232 KFO14640.1 KK696997 KFQ24952.1 KK501769 KFP17635.1 AGTO01000332 ABQF01011049 PZQS01000002 PVD35887.1 KL216593 KFV70296.1 KL460806 KFV12820.1 KK590766 KFV99933.1 HADW01020168 SBP21568.1 AADN05000350 HADX01006746 SBP28978.1 KK690812 KFQ16566.1 GBEX01002226 JAI12334.1 KK435974 KFQ90244.1 KK820088 KFQ39737.1 KK932664 KFQ45770.1 KK719330 KFO62656.1 JARO02008923 KPP62163.1 ADON01058539 ADON01058540 AERX01013232 AAGW02013744 AAGW02013745 AAGW02013746 AAGW02013747 AAGW02013748 AAGW02013749 AAGW02013750 KL425962 KFW90283.1 JH816117 EKC33010.1 FR904394 CDQ61999.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000235965
+ More
UP000002358 UP000215335 UP000000311 UP000007266 UP000076502 UP000005203 UP000053097 UP000279307 UP000242457 UP000008237 UP000005205 UP000192223 UP000053105 UP000030742 UP000019118 UP000078540 UP000007755 UP000078541 UP000078542 UP000009046 UP000245037 UP000078492 UP000189705 UP000085678 UP000076858 UP000261440 UP000053872 UP000190648 UP000054308 UP000000437 UP000018467 UP000007267 UP000053641 UP000053605 UP000054081 UP000053286 UP000053283 UP000053584 UP000053119 UP000016665 UP000007754 UP000245119 UP000053875 UP000000539 UP000261460 UP000261580 UP000265100 UP000265020 UP000001645 UP000052976 UP000264840 UP000261540 UP000034805 UP000016666 UP000005207 UP000001811 UP000005408 UP000193380
UP000002358 UP000215335 UP000000311 UP000007266 UP000076502 UP000005203 UP000053097 UP000279307 UP000242457 UP000008237 UP000005205 UP000192223 UP000053105 UP000030742 UP000019118 UP000078540 UP000007755 UP000078541 UP000078542 UP000009046 UP000245037 UP000078492 UP000189705 UP000085678 UP000076858 UP000261440 UP000053872 UP000190648 UP000054308 UP000000437 UP000018467 UP000007267 UP000053641 UP000053605 UP000054081 UP000053286 UP000053283 UP000053584 UP000053119 UP000016665 UP000007754 UP000245119 UP000053875 UP000000539 UP000261460 UP000261580 UP000265100 UP000265020 UP000001645 UP000052976 UP000264840 UP000261540 UP000034805 UP000016666 UP000005207 UP000001811 UP000005408 UP000193380
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IYR8
A0A2H1WMU9
A0A2A4JRU7
A0A212ET07
A0A0N0PEA9
A0A194QBK0
+ More
A0A2J7RK00 V5G7V2 K7ITX5 A0A232F0I9 E2ADZ0 D6W9D7 A0A154PJI0 A0A088A3A8 A0A026W4X0 A0A2A3E7H9 A0A0T6B6D3 E2BCE6 A0A0C9QGZ7 A0A310SL48 A0A158NBE2 A0A1W4WCU7 A0A0N0BCJ6 A0A1Y1KL99 U4U7A6 N6T9R4 A0A1B6J6D7 A0A195BAD5 E9IRA8 F4X8U7 A0A195FCQ1 A0A151IL05 E0VR31 A0A2P8ZGZ8 A0A195DRN1 A0A0C9RHA9 A0A1B6KH74 A0A0V0G930 A0A069DTZ1 A0A023F0T5 A0A1U7R678 A0A1U7RDE9 A0A1S3IHE9 A0A0P5K093 A0A0P5FVX3 A0A3B4DSY9 A0A2I0MG79 W5ULJ2 A0A0A9W7E4 A0A091MBQ8 A0A1V4K5Z3 A0A091I5C9 B0S5M0 R0LM96 W5KF39 K7F6Y1 A0A099YX94 Q58EC1 A0A091VHJ0 A0A093PAK9 A0A087QNP4 A0A091M0W4 A0A091GY55 A0A091UL85 A0A094K8J8 A0A093JVH6 A0A0A9YCE9 A0A087VPU8 A0A091QG71 A0A091JD49 U3KGG0 H0Z0N2 A0A2T7PR86 A0A093IUE0 A0A093CA41 A0A093IMF1 A0A1A7XU72 A0A3Q2U656 A0A3B4GUN0 A0A3Q4HLC0 A0A3P8Q918 A0A3Q2G300 A0A1A7YFH9 A0A091QAI3 G1NHV5 E1BQF6 A0A0F7ZCN0 A0A091UHJ8 A0A091RKD6 A0A091RSY9 A0A091F098 A0A3Q2UUK5 A0A3B3T8K6 A0A0P7U4Q6 U3INH1 I3J8N9 G1T9G4 A0A093QN06 K1QVP1 A0A060W9R2
A0A2J7RK00 V5G7V2 K7ITX5 A0A232F0I9 E2ADZ0 D6W9D7 A0A154PJI0 A0A088A3A8 A0A026W4X0 A0A2A3E7H9 A0A0T6B6D3 E2BCE6 A0A0C9QGZ7 A0A310SL48 A0A158NBE2 A0A1W4WCU7 A0A0N0BCJ6 A0A1Y1KL99 U4U7A6 N6T9R4 A0A1B6J6D7 A0A195BAD5 E9IRA8 F4X8U7 A0A195FCQ1 A0A151IL05 E0VR31 A0A2P8ZGZ8 A0A195DRN1 A0A0C9RHA9 A0A1B6KH74 A0A0V0G930 A0A069DTZ1 A0A023F0T5 A0A1U7R678 A0A1U7RDE9 A0A1S3IHE9 A0A0P5K093 A0A0P5FVX3 A0A3B4DSY9 A0A2I0MG79 W5ULJ2 A0A0A9W7E4 A0A091MBQ8 A0A1V4K5Z3 A0A091I5C9 B0S5M0 R0LM96 W5KF39 K7F6Y1 A0A099YX94 Q58EC1 A0A091VHJ0 A0A093PAK9 A0A087QNP4 A0A091M0W4 A0A091GY55 A0A091UL85 A0A094K8J8 A0A093JVH6 A0A0A9YCE9 A0A087VPU8 A0A091QG71 A0A091JD49 U3KGG0 H0Z0N2 A0A2T7PR86 A0A093IUE0 A0A093CA41 A0A093IMF1 A0A1A7XU72 A0A3Q2U656 A0A3B4GUN0 A0A3Q4HLC0 A0A3P8Q918 A0A3Q2G300 A0A1A7YFH9 A0A091QAI3 G1NHV5 E1BQF6 A0A0F7ZCN0 A0A091UHJ8 A0A091RKD6 A0A091RSY9 A0A091F098 A0A3Q2UUK5 A0A3B3T8K6 A0A0P7U4Q6 U3INH1 I3J8N9 G1T9G4 A0A093QN06 K1QVP1 A0A060W9R2
PDB
5F0P
E-value=4.93613e-09,
Score=146
Ontologies
GO
PANTHER
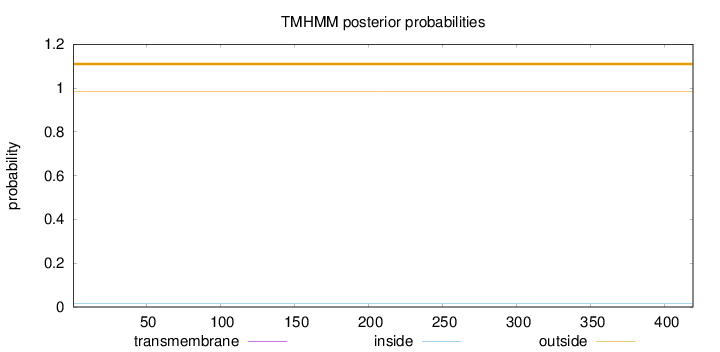
Topology
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00029
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01661
outside
1 - 419
Population Genetic Test Statistics
Pi
269.3546
Theta
187.428233
Tajima's D
1.549125
CLR
0.345202
CSRT
0.79726013699315
Interpretation
Uncertain
