Gene
KWMTBOMO05283
Pre Gene Modal
BGIBMGA002404
Annotation
PREDICTED:_cytosolic_carboxypeptidase-like_protein_5_isoform_X2_[Bombyx_mori]
Full name
Cytosolic carboxypeptidase-like protein 5
Alternative Name
ATP/GTP-binding protein-like 5
Location in the cell
Cytoplasmic Reliability : 1.011 Nuclear Reliability : 1.212
Sequence
CDS
ATGGATCAAAATAACATTATAGAATGTGGAGGATTTTATTTTATACATAATTTTGATTCTGGGAATCTGGGACATGTCGAAAGAGTACCGACAGAGCTTATAGCTCCTGGCATAAATCATAAAACAAATGCACCTGAAACACCAGACTATGAATTTAATATATGGACGCGTCCAGATTGTGCTGGTACCGAATTTGAGAATGGAAACCGAACCTGGTTCTACTTTGGTGTTCAAGCAACAGAGCAAAATGTTCTCGTGCGGTTAAACTTGGTAAACTTAAACAAACAAGGAAAGATGTACAATCAAGGTATGGCTCCAGTTACCAGAACCCTACCAGGCAAACCACAGTGGGAGAGGATCAGAGATAGACCCGTGCATTCGACCGACGACAACACATTCATGTTAAGTTTCAAATACAGAACGGCAGATAATTTGAAAGCAACGACATTCTTTGCCTTCACATATCCGTTCTCTTTTGCCGAACTACAGATAGCTTTGAACTCGATCGACCTAAAAATGCTACCTCTACCGCCCCCTCAGTCACCAGACGACATATACTACTATAGAGAATGTCTCATACATTCACTAGAAGGTAGGCGGGTGGATCTGCTAACTGTATCATCATACCACGGTATATCAACTGACAGGGAAGAAAAACTAACGAACCTCTTTCCGGAAAACCAAGAGAGGCCATTCAAGTTTCCTAATAAAAAGGTAGTGTTCATATCGGCGCGGGTACATCCAGGCGAGACGCCGTCCAGCTTCGTGTTCAATGGGTTTTTGAACCTACTCCTCACGAGGAACGATCCGGTCGCGATACAGTTGCGGAAACTATACGTCTTCAAGATGATACCGTTTCTGAATCCTGACGGAGTCGCCCGGGGCCACTACAGGACCGACACGAGGGGCGTTAATCTCAACAGAGTTTATTTAAATCCATCTCTGAGTTACCATCCAACGGTCTACGCTTCGAGGGCCTTGATAAGATATTATCACTTTGGTTGTGAAAAAGAAGACAGTTTCGAGGAGGGAAATCTATCCATGCGCAGCATCCAGAATGTCAACGAGACAAGCGAGACTTAG
Protein
MDQNNIIECGGFYFIHNFDSGNLGHVERVPTELIAPGINHKTNAPETPDYEFNIWTRPDCAGTEFENGNRTWFYFGVQATEQNVLVRLNLVNLNKQGKMYNQGMAPVTRTLPGKPQWERIRDRPVHSTDDNTFMLSFKYRTADNLKATTFFAFTYPFSFAELQIALNSIDLKMLPLPPPQSPDDIYYYRECLIHSLEGRRVDLLTVSSYHGISTDREEKLTNLFPENQERPFKFPNKKVVFISARVHPGETPSSFVFNGFLNLLLTRNDPVAIQLRKLYVFKMIPFLNPDGVARGHYRTDTRGVNLNRVYLNPSLSYHPTVYASRALIRYYHFGCEKEDSFEEGNLSMRSIQNVNETSET
Summary
Description
Metallocarboxypeptidase that mediates protein deglutamylation. Specifically catalyzes the deglutamylation of the branching point glutamate side chains generated by post-translational glutamylation in proteins such as tubulins. In contrast, it is not able to act as a long-chain deglutamylase that shortens long polyglutamate chains, a process catalyzed by AGTPBP1/CCP1, AGBL2/CCP2, AGBL3/CCP3, AGBL1/CCP4 and AGBL4/CCP6. Mediates deglutamylation of CGAS, regulating the antiviral activity of CGAS.
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M14 family.
Keywords
Alternative splicing
Carboxypeptidase
Complete proteome
Cytoplasm
Cytoskeleton
Disease mutation
Hydrolase
Metal-binding
Metalloprotease
Nucleus
Phosphoprotein
Polymorphism
Protease
Reference proteome
Retinitis pigmentosa
Zinc
Feature
chain Cytosolic carboxypeptidase-like protein 5
splice variant In isoform 3.
sequence variant In RP75; unknown pathological significance.
splice variant In isoform 3.
sequence variant In RP75; unknown pathological significance.
Uniprot
H9IYS0
A0A2H1WAK5
A0A212F1E3
A0A194QD67
A0A1E1W5B5
A0A0N1PJ56
+ More
D7EIR9 A0A0T6BCC1 A0A1Y1L9Z6 A0A1Y1L9X3 A0A2P8Z4J6 A0A1Y1L9V7 A0A1Y1LEP4 A0A1Y1LHF1 A0A1Y1L9X4 A0A1B6G9L3 A0A1B6IIP5 A0A1B6I206 A0A1B6KY10 A0A1B6M3L8 A0A2J7Q029 A0A2J7Q025 A0A348G602 A0A0L7RGR2 A0A088ADR5 A0A2A3EPE4 A0A1W4X7M2 A0A154PAK8 E1ZVU4 A0A0N0BBY4 F4WLT0 E0VU60 N6TUW6 A0A195FWC3 A0A151ITI0 A0A1B6E047 A0A1B6BZZ5 A0A151I4H5 A0A3Q0JHZ1 A0A232FA33 E2BW14 K7IZZ1 A0A026W388 A0A3L8DRZ6 M1ECV1 A0A3Q0IUK8 A0A067RR11 G1NVE8 A0A2F0BIA9 U6DM03 L5LLP7 S7N9X2 A0A2Y9ETM7 A0A0P6J6P0 A0A2G9RSI1 Q8NDL9-3 A0A384AVD9 U3DA09 A0A2J8VMS1 Q58CX9-2 H9EQT8 A0A2R9AVJ3 K7DJ08 G2HFJ2 A0A2Y9DN96 L8YBE8 M3YDF6 A0A2K6KUA3 F7BT54 A0A384AVE0 A0A3Q2I3I3 A0A384AVF0 A0A287ASG5 Q8NDL9-2 A0A2Y9NAI8 A0A341CZP9 A0A2J8VMS0 I3LU97 A0A2Y9N6P3 A0A286XJK3 H0WV03 A0A2I2YHG4 A0A3Q2LCQ5 A0A2K5QXN4 G3GXS1 A0A3Q1MYW4 G5BYZ0 L5KRI5 A0A0S7IFG7 A0A2Y9N5G1 A0A2Y9EUB7 A0A091CUV5 A0A2Y9ET83 A0A2J8NKZ2 G3TDG9 A0A2Y9EUY3 A0A212CY99 A0A3Q3EMS7 A0A2Y9NKX1 A0A2U4BZJ2
D7EIR9 A0A0T6BCC1 A0A1Y1L9Z6 A0A1Y1L9X3 A0A2P8Z4J6 A0A1Y1L9V7 A0A1Y1LEP4 A0A1Y1LHF1 A0A1Y1L9X4 A0A1B6G9L3 A0A1B6IIP5 A0A1B6I206 A0A1B6KY10 A0A1B6M3L8 A0A2J7Q029 A0A2J7Q025 A0A348G602 A0A0L7RGR2 A0A088ADR5 A0A2A3EPE4 A0A1W4X7M2 A0A154PAK8 E1ZVU4 A0A0N0BBY4 F4WLT0 E0VU60 N6TUW6 A0A195FWC3 A0A151ITI0 A0A1B6E047 A0A1B6BZZ5 A0A151I4H5 A0A3Q0JHZ1 A0A232FA33 E2BW14 K7IZZ1 A0A026W388 A0A3L8DRZ6 M1ECV1 A0A3Q0IUK8 A0A067RR11 G1NVE8 A0A2F0BIA9 U6DM03 L5LLP7 S7N9X2 A0A2Y9ETM7 A0A0P6J6P0 A0A2G9RSI1 Q8NDL9-3 A0A384AVD9 U3DA09 A0A2J8VMS1 Q58CX9-2 H9EQT8 A0A2R9AVJ3 K7DJ08 G2HFJ2 A0A2Y9DN96 L8YBE8 M3YDF6 A0A2K6KUA3 F7BT54 A0A384AVE0 A0A3Q2I3I3 A0A384AVF0 A0A287ASG5 Q8NDL9-2 A0A2Y9NAI8 A0A341CZP9 A0A2J8VMS0 I3LU97 A0A2Y9N6P3 A0A286XJK3 H0WV03 A0A2I2YHG4 A0A3Q2LCQ5 A0A2K5QXN4 G3GXS1 A0A3Q1MYW4 G5BYZ0 L5KRI5 A0A0S7IFG7 A0A2Y9N5G1 A0A2Y9EUB7 A0A091CUV5 A0A2Y9ET83 A0A2J8NKZ2 G3TDG9 A0A2Y9EUY3 A0A212CY99 A0A3Q3EMS7 A0A2Y9NKX1 A0A2U4BZJ2
EC Number
3.4.17.-
Pubmed
19121390
22118469
26354079
18362917
19820115
28004739
+ More
29403074 20798317 21719571 20566863 23537049 28648823 20075255 24508170 30249741 23236062 24845553 21993624 17974005 14702039 15815621 15489334 23085998 26720455 26355662 27842159 27764769 25243066 16305752 19390049 25319552 22722832 16136131 21484476 23385571 19892987 30723633 22398555 21804562 19393038 21993625 23258410
29403074 20798317 21719571 20566863 23537049 28648823 20075255 24508170 30249741 23236062 24845553 21993624 17974005 14702039 15815621 15489334 23085998 26720455 26355662 27842159 27764769 25243066 16305752 19390049 25319552 22722832 16136131 21484476 23385571 19892987 30723633 22398555 21804562 19393038 21993625 23258410
EMBL
BABH01010910
BABH01010911
BABH01010912
ODYU01007394
SOQ50111.1
AGBW02010898
+ More
OWR47541.1 KQ459220 KPJ02920.1 GDQN01008900 JAT82154.1 KQ459927 KPJ19309.1 DS497685 EFA12270.1 LJIG01002097 KRT84843.1 GEZM01061471 GEZM01061469 GEZM01061468 JAV70454.1 GEZM01061467 GEZM01061463 GEZM01061459 JAV70459.1 PYGN01000199 PSN51416.1 GEZM01061480 GEZM01061474 GEZM01061472 GEZM01061460 JAV70442.1 GEZM01061482 GEZM01061477 GEZM01061461 GEZM01061458 GEZM01061456 JAV70445.1 GEZM01061479 GEZM01061478 GEZM01061475 GEZM01061466 GEZM01061465 GEZM01061462 JAV70457.1 GEZM01061481 GEZM01061476 GEZM01061473 GEZM01061470 GEZM01061464 GEZM01061457 JAV70452.1 GECZ01010649 JAS59120.1 GECU01034832 GECU01020914 JAS72874.1 JAS86792.1 GECU01026750 JAS80956.1 GEBQ01023639 JAT16338.1 GEBQ01009489 GEBQ01004605 JAT30488.1 JAT35372.1 NEVH01020327 PNF21934.1 PNF21933.1 FX985541 BBF97875.1 KQ414596 KOC70019.1 KZ288206 PBC33076.1 KQ434864 KZC08976.1 GL434640 EFN74695.1 KQ435944 KOX68218.1 GL888216 EGI64812.1 DS235780 EEB16916.1 APGK01017572 APGK01017573 KB740042 ENN81868.1 KQ981204 KYN44965.1 KQ981015 KYN10277.1 GEDC01006000 JAS31298.1 GEDC01030461 JAS06837.1 KQ976462 KYM84654.1 NNAY01000597 OXU27482.1 GL451091 EFN80046.1 KK107510 EZA49504.1 QOIP01000004 RLU23205.1 JP005284 AER93881.1 KK852517 KDR22169.1 AAPE02039345 NTJE010217487 MBW02730.1 HAAF01011318 CCP83142.1 KB110330 ELK27389.1 KE163671 EPQ13100.1 GEBF01003736 JAN99896.1 KV931187 PIO30852.1 AL833844 AK302091 AC013403 AC013472 CH471053 BC007415 BC018584 BC131498 AK023398 AK025492 GAMT01005296 GAMS01006235 GAMR01008472 GAMQ01007201 GAMP01007223 JAB06565.1 JAB16901.1 JAB25460.1 JAB34650.1 JAB45532.1 NDHI03003415 PNJ58825.1 PNJ58826.1 PNJ58830.1 PNJ58832.1 BT021818 AAFC03088657 BC126504 JU320991 AFE64747.1 AJFE02095132 AJFE02095133 AJFE02095134 AACZ04002055 AACZ04002056 AACZ04002057 GABC01006352 GABF01009296 GABD01001683 GABE01002850 NBAG03000227 JAA04986.1 JAA12849.1 JAA31417.1 JAA41889.1 PNI72444.1 AK305506 BAK62500.1 KB360584 ELV13748.1 AEYP01063333 AEYP01063334 AEMK02000021 DQIR01065488 DQIR01068879 DQIR01071098 DQIR01085500 DQIR01098474 DQIR01098476 DQIR01161607 DQIR01167229 HDA20964.1 HDA53950.1 PNJ58828.1 DQIR01204849 HDB60326.1 AAKN02023455 AAQR03049341 AAQR03049342 CABD030013741 CABD030013742 CABD030013743 JH000061 EGV95860.1 JH172504 EHB14501.1 KB030580 ELK14054.1 GBYX01429443 JAO51848.1 KN123775 KFO23354.1 PNI72439.1 PNI72440.1 PNI72441.1 MKHE01000011 OWK10834.1
OWR47541.1 KQ459220 KPJ02920.1 GDQN01008900 JAT82154.1 KQ459927 KPJ19309.1 DS497685 EFA12270.1 LJIG01002097 KRT84843.1 GEZM01061471 GEZM01061469 GEZM01061468 JAV70454.1 GEZM01061467 GEZM01061463 GEZM01061459 JAV70459.1 PYGN01000199 PSN51416.1 GEZM01061480 GEZM01061474 GEZM01061472 GEZM01061460 JAV70442.1 GEZM01061482 GEZM01061477 GEZM01061461 GEZM01061458 GEZM01061456 JAV70445.1 GEZM01061479 GEZM01061478 GEZM01061475 GEZM01061466 GEZM01061465 GEZM01061462 JAV70457.1 GEZM01061481 GEZM01061476 GEZM01061473 GEZM01061470 GEZM01061464 GEZM01061457 JAV70452.1 GECZ01010649 JAS59120.1 GECU01034832 GECU01020914 JAS72874.1 JAS86792.1 GECU01026750 JAS80956.1 GEBQ01023639 JAT16338.1 GEBQ01009489 GEBQ01004605 JAT30488.1 JAT35372.1 NEVH01020327 PNF21934.1 PNF21933.1 FX985541 BBF97875.1 KQ414596 KOC70019.1 KZ288206 PBC33076.1 KQ434864 KZC08976.1 GL434640 EFN74695.1 KQ435944 KOX68218.1 GL888216 EGI64812.1 DS235780 EEB16916.1 APGK01017572 APGK01017573 KB740042 ENN81868.1 KQ981204 KYN44965.1 KQ981015 KYN10277.1 GEDC01006000 JAS31298.1 GEDC01030461 JAS06837.1 KQ976462 KYM84654.1 NNAY01000597 OXU27482.1 GL451091 EFN80046.1 KK107510 EZA49504.1 QOIP01000004 RLU23205.1 JP005284 AER93881.1 KK852517 KDR22169.1 AAPE02039345 NTJE010217487 MBW02730.1 HAAF01011318 CCP83142.1 KB110330 ELK27389.1 KE163671 EPQ13100.1 GEBF01003736 JAN99896.1 KV931187 PIO30852.1 AL833844 AK302091 AC013403 AC013472 CH471053 BC007415 BC018584 BC131498 AK023398 AK025492 GAMT01005296 GAMS01006235 GAMR01008472 GAMQ01007201 GAMP01007223 JAB06565.1 JAB16901.1 JAB25460.1 JAB34650.1 JAB45532.1 NDHI03003415 PNJ58825.1 PNJ58826.1 PNJ58830.1 PNJ58832.1 BT021818 AAFC03088657 BC126504 JU320991 AFE64747.1 AJFE02095132 AJFE02095133 AJFE02095134 AACZ04002055 AACZ04002056 AACZ04002057 GABC01006352 GABF01009296 GABD01001683 GABE01002850 NBAG03000227 JAA04986.1 JAA12849.1 JAA31417.1 JAA41889.1 PNI72444.1 AK305506 BAK62500.1 KB360584 ELV13748.1 AEYP01063333 AEYP01063334 AEMK02000021 DQIR01065488 DQIR01068879 DQIR01071098 DQIR01085500 DQIR01098474 DQIR01098476 DQIR01161607 DQIR01167229 HDA20964.1 HDA53950.1 PNJ58828.1 DQIR01204849 HDB60326.1 AAKN02023455 AAQR03049341 AAQR03049342 CABD030013741 CABD030013742 CABD030013743 JH000061 EGV95860.1 JH172504 EHB14501.1 KB030580 ELK14054.1 GBYX01429443 JAO51848.1 KN123775 KFO23354.1 PNI72439.1 PNI72440.1 PNI72441.1 MKHE01000011 OWK10834.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000007266
UP000245037
+ More
UP000235965 UP000053825 UP000005203 UP000242457 UP000192223 UP000076502 UP000000311 UP000053105 UP000007755 UP000009046 UP000019118 UP000078541 UP000078492 UP000078540 UP000079169 UP000215335 UP000008237 UP000002358 UP000053097 UP000279307 UP000027135 UP000001074 UP000248484 UP000005640 UP000261681 UP000009136 UP000240080 UP000002277 UP000248480 UP000011518 UP000000715 UP000233180 UP000002281 UP000008227 UP000248483 UP000252040 UP000005447 UP000005225 UP000001519 UP000233040 UP000001075 UP000006813 UP000010552 UP000028990 UP000007646 UP000261660 UP000245320
UP000235965 UP000053825 UP000005203 UP000242457 UP000192223 UP000076502 UP000000311 UP000053105 UP000007755 UP000009046 UP000019118 UP000078541 UP000078492 UP000078540 UP000079169 UP000215335 UP000008237 UP000002358 UP000053097 UP000279307 UP000027135 UP000001074 UP000248484 UP000005640 UP000261681 UP000009136 UP000240080 UP000002277 UP000248480 UP000011518 UP000000715 UP000233180 UP000002281 UP000008227 UP000248483 UP000252040 UP000005447 UP000005225 UP000001519 UP000233040 UP000001075 UP000006813 UP000010552 UP000028990 UP000007646 UP000261660 UP000245320
ProteinModelPortal
H9IYS0
A0A2H1WAK5
A0A212F1E3
A0A194QD67
A0A1E1W5B5
A0A0N1PJ56
+ More
D7EIR9 A0A0T6BCC1 A0A1Y1L9Z6 A0A1Y1L9X3 A0A2P8Z4J6 A0A1Y1L9V7 A0A1Y1LEP4 A0A1Y1LHF1 A0A1Y1L9X4 A0A1B6G9L3 A0A1B6IIP5 A0A1B6I206 A0A1B6KY10 A0A1B6M3L8 A0A2J7Q029 A0A2J7Q025 A0A348G602 A0A0L7RGR2 A0A088ADR5 A0A2A3EPE4 A0A1W4X7M2 A0A154PAK8 E1ZVU4 A0A0N0BBY4 F4WLT0 E0VU60 N6TUW6 A0A195FWC3 A0A151ITI0 A0A1B6E047 A0A1B6BZZ5 A0A151I4H5 A0A3Q0JHZ1 A0A232FA33 E2BW14 K7IZZ1 A0A026W388 A0A3L8DRZ6 M1ECV1 A0A3Q0IUK8 A0A067RR11 G1NVE8 A0A2F0BIA9 U6DM03 L5LLP7 S7N9X2 A0A2Y9ETM7 A0A0P6J6P0 A0A2G9RSI1 Q8NDL9-3 A0A384AVD9 U3DA09 A0A2J8VMS1 Q58CX9-2 H9EQT8 A0A2R9AVJ3 K7DJ08 G2HFJ2 A0A2Y9DN96 L8YBE8 M3YDF6 A0A2K6KUA3 F7BT54 A0A384AVE0 A0A3Q2I3I3 A0A384AVF0 A0A287ASG5 Q8NDL9-2 A0A2Y9NAI8 A0A341CZP9 A0A2J8VMS0 I3LU97 A0A2Y9N6P3 A0A286XJK3 H0WV03 A0A2I2YHG4 A0A3Q2LCQ5 A0A2K5QXN4 G3GXS1 A0A3Q1MYW4 G5BYZ0 L5KRI5 A0A0S7IFG7 A0A2Y9N5G1 A0A2Y9EUB7 A0A091CUV5 A0A2Y9ET83 A0A2J8NKZ2 G3TDG9 A0A2Y9EUY3 A0A212CY99 A0A3Q3EMS7 A0A2Y9NKX1 A0A2U4BZJ2
D7EIR9 A0A0T6BCC1 A0A1Y1L9Z6 A0A1Y1L9X3 A0A2P8Z4J6 A0A1Y1L9V7 A0A1Y1LEP4 A0A1Y1LHF1 A0A1Y1L9X4 A0A1B6G9L3 A0A1B6IIP5 A0A1B6I206 A0A1B6KY10 A0A1B6M3L8 A0A2J7Q029 A0A2J7Q025 A0A348G602 A0A0L7RGR2 A0A088ADR5 A0A2A3EPE4 A0A1W4X7M2 A0A154PAK8 E1ZVU4 A0A0N0BBY4 F4WLT0 E0VU60 N6TUW6 A0A195FWC3 A0A151ITI0 A0A1B6E047 A0A1B6BZZ5 A0A151I4H5 A0A3Q0JHZ1 A0A232FA33 E2BW14 K7IZZ1 A0A026W388 A0A3L8DRZ6 M1ECV1 A0A3Q0IUK8 A0A067RR11 G1NVE8 A0A2F0BIA9 U6DM03 L5LLP7 S7N9X2 A0A2Y9ETM7 A0A0P6J6P0 A0A2G9RSI1 Q8NDL9-3 A0A384AVD9 U3DA09 A0A2J8VMS1 Q58CX9-2 H9EQT8 A0A2R9AVJ3 K7DJ08 G2HFJ2 A0A2Y9DN96 L8YBE8 M3YDF6 A0A2K6KUA3 F7BT54 A0A384AVE0 A0A3Q2I3I3 A0A384AVF0 A0A287ASG5 Q8NDL9-2 A0A2Y9NAI8 A0A341CZP9 A0A2J8VMS0 I3LU97 A0A2Y9N6P3 A0A286XJK3 H0WV03 A0A2I2YHG4 A0A3Q2LCQ5 A0A2K5QXN4 G3GXS1 A0A3Q1MYW4 G5BYZ0 L5KRI5 A0A0S7IFG7 A0A2Y9N5G1 A0A2Y9EUB7 A0A091CUV5 A0A2Y9ET83 A0A2J8NKZ2 G3TDG9 A0A2Y9EUY3 A0A212CY99 A0A3Q3EMS7 A0A2Y9NKX1 A0A2U4BZJ2
PDB
3L2N
E-value=7.67674e-16,
Score=204
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Cytosol Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Nucleus Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Cytoskeleton Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Spindle Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Midbody Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Cytosol Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Nucleus Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Cytoskeleton Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Spindle Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
Midbody Mainly cytoplasmic. Slight accumulation in the nucleus is observed (By similarity). Colocalizes with alpha-tubulin in the mitotic spindle and with midbody microtubules in the intercellular bridges formed during cytokinesis. With evidence from 2 publications.
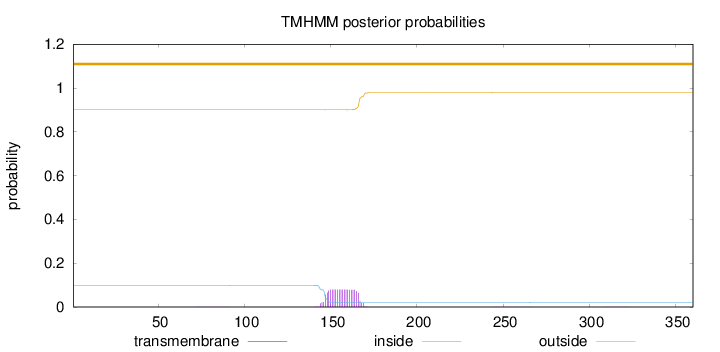
Length:
360
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.67093
Exp number, first 60 AAs:
0.00182
Total prob of N-in:
0.09898
outside
1 - 360
Population Genetic Test Statistics
Pi
26.900749
Theta
23.898301
Tajima's D
0.3789
CLR
0.018253
CSRT
0.484475776211189
Interpretation
Uncertain
