Pre Gene Modal
BGIBMGA002504
Annotation
PREDICTED:_GTP-binding_protein_SAR1b_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.667
Sequence
CDS
ATGTTTATCCTCGACTGGTTCACTGGAGTGTTGGGATTTCTTGGTCTATGGAAAAAGTCGGGCAAGCTCTTGTTTCTGGGACTTGACAATGCAGGGAAGACAACACTCTTACATATGTTGAAAGATGATAGACTTGCTCAACATGTACCTACTTTACATCCTACATCCGAGGAACTGTCAATAGGCAGTATGCGTTTCACGACGTTCGACCTCGGTGGGCATCAGCAGGCGCGTCGTGTGTGGCGCGACTACTTCCCCGCGGTGGACGCGATAGTGTTCCTGGTCGACGCGTGTGACCGGCCACGTCTGCCGGAGTCCAAGGCTGAACTAGACTCGTTGCTCACTGACGAGACGCTCAGTAACTGCCCTGTGCTGATTCTCGGCAACAAGATTGATAAACCCGGTGCTGCCAGTGAAGACGAACTACGTCACTTTTTCAATCTGTATCAGCAGACCACTGGAAAGGGAAAAGTATCAAGGTCCGAGCTTCCCGGTCGGCCGCTGGAGCTTTTCATGTGCTCGGTGCTGAAGCGTCAGGGCTACGGCGAGGGCTTCCGCTGGCTCGCGCAGTACATCGACTGA
Protein
MFILDWFTGVLGFLGLWKKSGKLLFLGLDNAGKTTLLHMLKDDRLAQHVPTLHPTSEELSIGSMRFTTFDLGGHQQARRVWRDYFPAVDAIVFLVDACDRPRLPESKAELDSLLTDETLSNCPVLILGNKIDKPGAASEDELRHFFNLYQQTTGKGKVSRSELPGRPLELFMCSVLKRQGYGEGFRWLAQYID
Summary
Similarity
Belongs to the small GTPase superfamily. SAR1 family.
Uniprot
H9IZ19
A0A2H1WAN2
A0A2A4IYR9
A0A0N1PHE7
I4DN84
I4DRE7
+ More
S4P8J5 A0A212F1F4 H9KQA6 A0A084VCI0 A0A1L8DVR0 A0A0K8TQ80 K7IWW9 T1E7J6 A0A2M3ZFJ5 A0A2M4A582 W5JFN0 E9IZM4 F4WDI7 A0A0J7NLG9 A0A182MT63 A0A154P462 A0A158NIP5 A0A182IKC5 E2BCN9 A0A182RID1 A0A182PNM6 A0A182W0G5 A0A182N5K6 T1D4Y7 E0W2R7 E2A6R9 A0A182SR63 Q1HR30 A0A1Q3FFA1 B0WY19 A0A182HCE3 U5EL87 B7UEA5 B4K729 B4M0D6 A0A026WM88 R4WQP7 B4NHT7 A0A1W4W2J6 A0A182V0S7 A0A182JSH6 A0A182UH17 A0A182XFJ7 Q7PQL9 A0A182I7Z9 B4JRN9 D1FPP4 A0A2A3E438 A0A0M4EQT2 A0A0L0BZ28 T1P8X1 A0A1I8NY32 A0A2I9LPQ4 B4QZZ3 Q298N7 B4HE63 B3LVN7 Q9VD29 B4PLM8 B3P7K9 A0A0L7R4B6 A0A165VFS5 A0A034WFM1 A0A0K8VFE4 W8C233 A0A1L8EH40 A0A3B0K9N0 B4G4D3 A0A0P4VML0 R4G581 A0A1A9W2U5 A0A2J7Q2X0 A0A023F9C0 A0A1Z5L244 A0A310SN13 A0A226EPZ0 A0A293LYK4 A0A182F775 A0A2R5LDF5 A0A1W7R9L9 A0A224XL52 A0A069DQD2 A0A1Z5L7D3 A0A131XUP2 B7PDY5 L7M4P7 A0A0A9XJM6 U4UP32 D3TM22 A0A1A9YFT2 A0A1A9VLW0 A0A1B0AV77 A0A1A9ZQ43 C1BQW8
S4P8J5 A0A212F1F4 H9KQA6 A0A084VCI0 A0A1L8DVR0 A0A0K8TQ80 K7IWW9 T1E7J6 A0A2M3ZFJ5 A0A2M4A582 W5JFN0 E9IZM4 F4WDI7 A0A0J7NLG9 A0A182MT63 A0A154P462 A0A158NIP5 A0A182IKC5 E2BCN9 A0A182RID1 A0A182PNM6 A0A182W0G5 A0A182N5K6 T1D4Y7 E0W2R7 E2A6R9 A0A182SR63 Q1HR30 A0A1Q3FFA1 B0WY19 A0A182HCE3 U5EL87 B7UEA5 B4K729 B4M0D6 A0A026WM88 R4WQP7 B4NHT7 A0A1W4W2J6 A0A182V0S7 A0A182JSH6 A0A182UH17 A0A182XFJ7 Q7PQL9 A0A182I7Z9 B4JRN9 D1FPP4 A0A2A3E438 A0A0M4EQT2 A0A0L0BZ28 T1P8X1 A0A1I8NY32 A0A2I9LPQ4 B4QZZ3 Q298N7 B4HE63 B3LVN7 Q9VD29 B4PLM8 B3P7K9 A0A0L7R4B6 A0A165VFS5 A0A034WFM1 A0A0K8VFE4 W8C233 A0A1L8EH40 A0A3B0K9N0 B4G4D3 A0A0P4VML0 R4G581 A0A1A9W2U5 A0A2J7Q2X0 A0A023F9C0 A0A1Z5L244 A0A310SN13 A0A226EPZ0 A0A293LYK4 A0A182F775 A0A2R5LDF5 A0A1W7R9L9 A0A224XL52 A0A069DQD2 A0A1Z5L7D3 A0A131XUP2 B7PDY5 L7M4P7 A0A0A9XJM6 U4UP32 D3TM22 A0A1A9YFT2 A0A1A9VLW0 A0A1B0AV77 A0A1A9ZQ43 C1BQW8
Pubmed
19121390
26354079
22651552
23622113
22118469
24438588
+ More
26369729 20075255 20920257 23761445 21282665 21719571 21347285 20798317 24330624 20566863 17204158 17510324 26483478 17994087 18057021 24508170 30249741 23691247 12364791 14747013 17210077 20441151 26108605 25315136 29248469 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 24495485 27129103 25474469 28528879 30926861 26334808 25576852 25401762 26823975 23537049 20353571
26369729 20075255 20920257 23761445 21282665 21719571 21347285 20798317 24330624 20566863 17204158 17510324 26483478 17994087 18057021 24508170 30249741 23691247 12364791 14747013 17210077 20441151 26108605 25315136 29248469 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 24495485 27129103 25474469 28528879 30926861 26334808 25576852 25401762 26823975 23537049 20353571
EMBL
BABH01010909
ODYU01007394
SOQ50113.1
NWSH01004579
PCG64901.1
KQ459927
+ More
KPJ19306.1 AK402769 KQ459220 BAM19374.1 KPJ02918.1 AK405086 BAM20487.1 GAIX01005901 JAA86659.1 AGBW02010898 OWR47543.1 ATLV01010759 KE524612 KFB35674.1 GFDF01003566 JAV10518.1 GDAI01001300 JAI16303.1 GAMD01002982 JAA98608.1 GGFM01006586 MBW27337.1 GGFK01002608 MBW35929.1 ADMH02001534 GGFL01004218 ETN62148.1 MBW68396.1 GL767232 EFZ13957.1 GL888087 EGI67767.1 LBMM01003615 KMQ93320.1 AXCM01002248 KQ434810 KZC06739.1 ADTU01017041 GL447336 EFN86549.1 GALA01000773 JAA94079.1 DS235879 EEB19923.1 GL437197 EFN70863.1 DQ440264 CH477631 ABF18297.1 EAT38070.1 GFDL01008818 JAV26227.1 DS232179 EDS36829.1 JXUM01127485 JXUM01127486 JXUM01127487 GANO01001511 JAB58360.1 FJ538185 ACK44330.1 CH933806 EDW16342.1 KRG02054.1 KRG02055.1 KRG02056.1 KRG02057.1 CH940650 EDW67298.1 KRF83231.1 KK107152 QOIP01000007 EZA57172.1 RLU20109.1 AK417002 BAN20217.1 CH964272 EDW84697.1 AAAB01008880 EAA08621.2 EGK96987.1 APCN01002591 CH916373 EDV94429.1 EZ419794 ACY69967.1 KZ288383 PBC26497.1 CP012526 ALC47414.1 JRES01001133 KNC25265.1 KA644590 AFP59219.1 GFWZ01000375 MBW20365.1 CM000364 EDX13899.1 CM000070 EAL27918.1 CH480815 EDW43160.1 CH902617 EDV42607.1 KPU79813.1 AE014297 BT001745 AAF55974.1 AAN14369.1 AAN14370.1 AAN71500.1 AAS65194.1 CM000160 EDW97977.1 KRK04043.1 CH954182 EDV54170.1 KQ414657 KOC65722.1 KT984793 AMZ00345.1 GAKP01004591 JAC54361.1 GDHF01032176 GDHF01025062 GDHF01014708 GDHF01004886 JAI20138.1 JAI27252.1 JAI37606.1 JAI47428.1 GAMC01000668 GAMC01000667 JAC05888.1 GFDG01000749 JAV18050.1 OUUW01000007 SPP82779.1 CH479179 EDW24481.1 GDKW01000807 JAI55788.1 ACPB03002694 GAHY01000740 JAA76770.1 NEVH01019075 PNF22935.1 GBBI01000616 JAC18096.1 GFJQ02005600 JAW01370.1 KQ761799 OAD56783.1 LNIX01000002 MH602479 OXA59559.1 QBH72724.1 GFWV01009064 MAA33793.1 GGLE01003434 MBY07560.1 GFAH01000550 JAV47839.1 GFTR01003210 JAW13216.1 GBGD01003062 JAC85827.1 GFJQ02004006 JAW02964.1 GEFM01005419 JAP70377.1 ABJB010032831 ABJB010221700 ABJB010226926 ABJB010732850 DS692829 EEC04807.1 GACK01006019 JAA59015.1 GBHO01026294 GBRD01011954 GDHC01020482 JAG17310.1 JAG53870.1 JAP98146.1 KB632306 ERL91901.1 EZ422474 ADD18750.1 JXJN01004067 BT076997 ACO11421.1
KPJ19306.1 AK402769 KQ459220 BAM19374.1 KPJ02918.1 AK405086 BAM20487.1 GAIX01005901 JAA86659.1 AGBW02010898 OWR47543.1 ATLV01010759 KE524612 KFB35674.1 GFDF01003566 JAV10518.1 GDAI01001300 JAI16303.1 GAMD01002982 JAA98608.1 GGFM01006586 MBW27337.1 GGFK01002608 MBW35929.1 ADMH02001534 GGFL01004218 ETN62148.1 MBW68396.1 GL767232 EFZ13957.1 GL888087 EGI67767.1 LBMM01003615 KMQ93320.1 AXCM01002248 KQ434810 KZC06739.1 ADTU01017041 GL447336 EFN86549.1 GALA01000773 JAA94079.1 DS235879 EEB19923.1 GL437197 EFN70863.1 DQ440264 CH477631 ABF18297.1 EAT38070.1 GFDL01008818 JAV26227.1 DS232179 EDS36829.1 JXUM01127485 JXUM01127486 JXUM01127487 GANO01001511 JAB58360.1 FJ538185 ACK44330.1 CH933806 EDW16342.1 KRG02054.1 KRG02055.1 KRG02056.1 KRG02057.1 CH940650 EDW67298.1 KRF83231.1 KK107152 QOIP01000007 EZA57172.1 RLU20109.1 AK417002 BAN20217.1 CH964272 EDW84697.1 AAAB01008880 EAA08621.2 EGK96987.1 APCN01002591 CH916373 EDV94429.1 EZ419794 ACY69967.1 KZ288383 PBC26497.1 CP012526 ALC47414.1 JRES01001133 KNC25265.1 KA644590 AFP59219.1 GFWZ01000375 MBW20365.1 CM000364 EDX13899.1 CM000070 EAL27918.1 CH480815 EDW43160.1 CH902617 EDV42607.1 KPU79813.1 AE014297 BT001745 AAF55974.1 AAN14369.1 AAN14370.1 AAN71500.1 AAS65194.1 CM000160 EDW97977.1 KRK04043.1 CH954182 EDV54170.1 KQ414657 KOC65722.1 KT984793 AMZ00345.1 GAKP01004591 JAC54361.1 GDHF01032176 GDHF01025062 GDHF01014708 GDHF01004886 JAI20138.1 JAI27252.1 JAI37606.1 JAI47428.1 GAMC01000668 GAMC01000667 JAC05888.1 GFDG01000749 JAV18050.1 OUUW01000007 SPP82779.1 CH479179 EDW24481.1 GDKW01000807 JAI55788.1 ACPB03002694 GAHY01000740 JAA76770.1 NEVH01019075 PNF22935.1 GBBI01000616 JAC18096.1 GFJQ02005600 JAW01370.1 KQ761799 OAD56783.1 LNIX01000002 MH602479 OXA59559.1 QBH72724.1 GFWV01009064 MAA33793.1 GGLE01003434 MBY07560.1 GFAH01000550 JAV47839.1 GFTR01003210 JAW13216.1 GBGD01003062 JAC85827.1 GFJQ02004006 JAW02964.1 GEFM01005419 JAP70377.1 ABJB010032831 ABJB010221700 ABJB010226926 ABJB010732850 DS692829 EEC04807.1 GACK01006019 JAA59015.1 GBHO01026294 GBRD01011954 GDHC01020482 JAG17310.1 JAG53870.1 JAP98146.1 KB632306 ERL91901.1 EZ422474 ADD18750.1 JXJN01004067 BT076997 ACO11421.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000005203
+ More
UP000030765 UP000002358 UP000000673 UP000007755 UP000036403 UP000075883 UP000076502 UP000005205 UP000075880 UP000008237 UP000075900 UP000075885 UP000075920 UP000075884 UP000009046 UP000000311 UP000075901 UP000008820 UP000002320 UP000069940 UP000009192 UP000008792 UP000053097 UP000279307 UP000007798 UP000192221 UP000075903 UP000075881 UP000075902 UP000076407 UP000007062 UP000075840 UP000001070 UP000242457 UP000092553 UP000037069 UP000095301 UP000095300 UP000000304 UP000001819 UP000001292 UP000007801 UP000000803 UP000002282 UP000008711 UP000053825 UP000268350 UP000008744 UP000015103 UP000091820 UP000235965 UP000198287 UP000069272 UP000001555 UP000030742 UP000092443 UP000078200 UP000092460 UP000092445
UP000030765 UP000002358 UP000000673 UP000007755 UP000036403 UP000075883 UP000076502 UP000005205 UP000075880 UP000008237 UP000075900 UP000075885 UP000075920 UP000075884 UP000009046 UP000000311 UP000075901 UP000008820 UP000002320 UP000069940 UP000009192 UP000008792 UP000053097 UP000279307 UP000007798 UP000192221 UP000075903 UP000075881 UP000075902 UP000076407 UP000007062 UP000075840 UP000001070 UP000242457 UP000092553 UP000037069 UP000095301 UP000095300 UP000000304 UP000001819 UP000001292 UP000007801 UP000000803 UP000002282 UP000008711 UP000053825 UP000268350 UP000008744 UP000015103 UP000091820 UP000235965 UP000198287 UP000069272 UP000001555 UP000030742 UP000092443 UP000078200 UP000092460 UP000092445
Pfam
PF00025 Arf
Interpro
ProteinModelPortal
H9IZ19
A0A2H1WAN2
A0A2A4IYR9
A0A0N1PHE7
I4DN84
I4DRE7
+ More
S4P8J5 A0A212F1F4 H9KQA6 A0A084VCI0 A0A1L8DVR0 A0A0K8TQ80 K7IWW9 T1E7J6 A0A2M3ZFJ5 A0A2M4A582 W5JFN0 E9IZM4 F4WDI7 A0A0J7NLG9 A0A182MT63 A0A154P462 A0A158NIP5 A0A182IKC5 E2BCN9 A0A182RID1 A0A182PNM6 A0A182W0G5 A0A182N5K6 T1D4Y7 E0W2R7 E2A6R9 A0A182SR63 Q1HR30 A0A1Q3FFA1 B0WY19 A0A182HCE3 U5EL87 B7UEA5 B4K729 B4M0D6 A0A026WM88 R4WQP7 B4NHT7 A0A1W4W2J6 A0A182V0S7 A0A182JSH6 A0A182UH17 A0A182XFJ7 Q7PQL9 A0A182I7Z9 B4JRN9 D1FPP4 A0A2A3E438 A0A0M4EQT2 A0A0L0BZ28 T1P8X1 A0A1I8NY32 A0A2I9LPQ4 B4QZZ3 Q298N7 B4HE63 B3LVN7 Q9VD29 B4PLM8 B3P7K9 A0A0L7R4B6 A0A165VFS5 A0A034WFM1 A0A0K8VFE4 W8C233 A0A1L8EH40 A0A3B0K9N0 B4G4D3 A0A0P4VML0 R4G581 A0A1A9W2U5 A0A2J7Q2X0 A0A023F9C0 A0A1Z5L244 A0A310SN13 A0A226EPZ0 A0A293LYK4 A0A182F775 A0A2R5LDF5 A0A1W7R9L9 A0A224XL52 A0A069DQD2 A0A1Z5L7D3 A0A131XUP2 B7PDY5 L7M4P7 A0A0A9XJM6 U4UP32 D3TM22 A0A1A9YFT2 A0A1A9VLW0 A0A1B0AV77 A0A1A9ZQ43 C1BQW8
S4P8J5 A0A212F1F4 H9KQA6 A0A084VCI0 A0A1L8DVR0 A0A0K8TQ80 K7IWW9 T1E7J6 A0A2M3ZFJ5 A0A2M4A582 W5JFN0 E9IZM4 F4WDI7 A0A0J7NLG9 A0A182MT63 A0A154P462 A0A158NIP5 A0A182IKC5 E2BCN9 A0A182RID1 A0A182PNM6 A0A182W0G5 A0A182N5K6 T1D4Y7 E0W2R7 E2A6R9 A0A182SR63 Q1HR30 A0A1Q3FFA1 B0WY19 A0A182HCE3 U5EL87 B7UEA5 B4K729 B4M0D6 A0A026WM88 R4WQP7 B4NHT7 A0A1W4W2J6 A0A182V0S7 A0A182JSH6 A0A182UH17 A0A182XFJ7 Q7PQL9 A0A182I7Z9 B4JRN9 D1FPP4 A0A2A3E438 A0A0M4EQT2 A0A0L0BZ28 T1P8X1 A0A1I8NY32 A0A2I9LPQ4 B4QZZ3 Q298N7 B4HE63 B3LVN7 Q9VD29 B4PLM8 B3P7K9 A0A0L7R4B6 A0A165VFS5 A0A034WFM1 A0A0K8VFE4 W8C233 A0A1L8EH40 A0A3B0K9N0 B4G4D3 A0A0P4VML0 R4G581 A0A1A9W2U5 A0A2J7Q2X0 A0A023F9C0 A0A1Z5L244 A0A310SN13 A0A226EPZ0 A0A293LYK4 A0A182F775 A0A2R5LDF5 A0A1W7R9L9 A0A224XL52 A0A069DQD2 A0A1Z5L7D3 A0A131XUP2 B7PDY5 L7M4P7 A0A0A9XJM6 U4UP32 D3TM22 A0A1A9YFT2 A0A1A9VLW0 A0A1B0AV77 A0A1A9ZQ43 C1BQW8
PDB
1F6B
E-value=1.70545e-83,
Score=784
Ontologies
GO
GO:0005794
GO:0016192
GO:0005783
GO:0006886
GO:0005525
GO:0050775
GO:0070863
GO:0000902
GO:0097110
GO:0072659
GO:0035050
GO:0035293
GO:0009306
GO:0070971
GO:0007029
GO:0048081
GO:1905808
GO:0030127
GO:0061024
GO:0016050
GO:0003924
GO:0003400
GO:0006888
GO:0008363
GO:0040003
GO:0005622
GO:0051920
GO:0003887
GO:0005524
GO:0001664
GO:0016491
GO:0003824
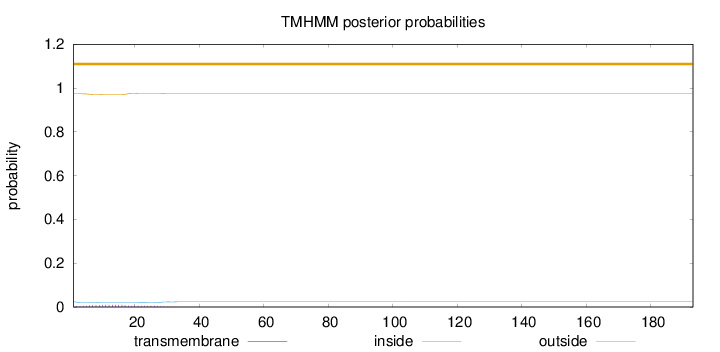
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19062
Exp number, first 60 AAs:
0.19026
Total prob of N-in:
0.02556
outside
1 - 193
Population Genetic Test Statistics
Pi
215.466677
Theta
200.285883
Tajima's D
0.206485
CLR
1.134951
CSRT
0.428778561071946
Interpretation
Uncertain
