Pre Gene Modal
BGIBMGA002502
Annotation
PREDICTED:_histone-lysine_N-methyltransferase_2D-like?_partial_[Bombyx_mori]
Full name
Histone-lysine N-methyltransferase
Location in the cell
Nuclear Reliability : 3.288
Sequence
CDS
ATGGAAGTACCGGACGAAAATGTTCTAGAGGTAGATTTGTTAGCTGATGATGGATCAGAAGATGAAGGCGAAGCCCCGCCGTCTTCAGAATTTTACACCGGACCTGGGTCGACACCAACATCATGTGCGTCATCTCCTCGAGGTGAAGAACCAGCATCTCCCCATGCAGGTCTCACACAACCATCCTTCTTCCATCATCAAGGTTACACTCGCCCGTTTTTTACCTCGGCCAGGCGAGGTCCGGGTAGACCACGGAAAGAAGGACCCAAGTTAGCTCGTGAAGGCAAGATTGTTAGAAGATACAGAGGAAATCCAGGATCTTTAAGAGGGGCAAAAAGACATCGTGGTAGTAGGGATGATGCATCTGAAGACATGATGGACGAAGATGATTTTACAATACCACCTGTCCCCGAGGAACCTCCCTATATGCCCGAAAAATGGCCTGGTAAAGTTTGCTCACTTTGTAACTTGGGTGAACGCAGTCAACTAGGCCAGGGTGAAATGAGACAAATTCATTGCAATATTGGTGAAGCAGAGGGTAGTACGACACCTTTAGTTACCAATTCAGGAGGAGCCACACCAACAAGCATTGCTACTCCTACTAGTACTCCAACGACACCTGTCACACCAGGCTTAGTTTCTCCTCCACCAGAGCTAGTAGATCCCAACCAACATCCTCTTGGGCTACCGCTCAGCAGGCGCCAGAAATCTTTCAACAAGTGCAAAACACCACTCTACAATATGGAACACACAGATGAATTATCAATTATCGGTCACAATGACTCTCTAGAAATACAAGCAGTAGTGTCTTCTGGAGCATTGTACATTCACCGTTGTTGCCTTGAGTTCAGTCCACCATTCCAAGCAACATCATCTGAGGAAGACCTTGAGCAAGCAGAGGAGACGAGAATCAGAGGAATAGTCACTTCTGCTCTTACAAGAAAATGTGCCTTTTGTACTAGACATGGTGCAAGTATTCCTTGTAAGATGAGCTGTAACAAATACTACCATCTACCATGTCTTCTAGCATCAGGTGGTTTTATGGATTTTCAAAGCAAGGGCTCATTTTGTAAAGATCATTTATATCAAGTTCCCCTTGTCTGTACATCAGAAATCGACTGCAGGACTTGTAGGACTATCGGCGATATTGCGAATCTGATGACTTGCGTGGTTTGTGGAGCGCATTATCATGGAACTTGTGTCGGTCTTGCTCAGCTACCAGGCGTCCGGTCGGGCTGGGCGTGCCGCGGGTGCCGCGTGTGCCAGGTGTGCCGCGAGCCGGCGCCGGGCGAGGCGCGCGCCGTGTGCTGCGACCACTGCGACAAGCTCTACCACGCCGCCTGTCTACGACCTCTCATGGCCACCGTGCCCAAGTACGGCTGGAAGTGCAAGTGTTGCCGGGTGTGCTCCGACTGCGGGTCGCGGTCGCCGGGCGCCGGGCCGTCGTCCCGCTGGCACGCGCACTACACGGTGTGCGACTCGTGCTACCAGCAGCGCAACAAGGGCTCGTGCTGTCCGCTGTGTCGCCGAGCTTACCGCGCCGCCGCCTACCGCGACATGATACGGTGCACGCTCTGCAAGAGATACGTCCATGGCACCTGTGATCCAGATGCGGAGCCTCAACAGTATAGGAAAAACAAAGATCAGAATCCATCTTATGAATACAGCTGTCCTATATGCAAGAGTCAACTTGCACTGACCGGATCTAAACCTGGGTCTTTTGAAGAAGATAGCACTGCTTCAGTGTCTCAAGATTCTTCATATGGAGATGACAACTCTAATTTACAAGATCAGGACCCACTTGCGATTGAAACAAAGCCTGATGTTGGTCTTGGAAAAGGAAAACCTTTCTCAGTATCTTCAAAGGTAGCCAAAAAAAAAATTGGAGGCTACAAACTGAAAGGTGGATTCCCTGCGGGTGGCAAGTTGGGCTTCCAGAAACGCCAACGTTCTCTGTTGGACTTCGGTCGGAAAAGGGCATCTAAACCTAAAATGCGAGGAGTTTTTGGAGTACCAGGATTGGGGCTACAGAGACCCCAAGCTCCTGACAGTAAAAGCTCTGAGGATGATCCAGGAATGGAGAATAAACTAGTGCTGTGTTCAAGCAAAGACAAGTTTGTGCTCACACAAGATTTGTGCGTTATGTGTGGTGCTGTCGGTACGGACTCTGAGGGATGCCTTATTGCTTGTGCCCAGTGTGGACAGACTTATCATCCGTACTGCGTAAACATAAAGGTGATACAAAATTAA
Protein
MEVPDENVLEVDLLADDGSEDEGEAPPSSEFYTGPGSTPTSCASSPRGEEPASPHAGLTQPSFFHHQGYTRPFFTSARRGPGRPRKEGPKLAREGKIVRRYRGNPGSLRGAKRHRGSRDDASEDMMDEDDFTIPPVPEEPPYMPEKWPGKVCSLCNLGERSQLGQGEMRQIHCNIGEAEGSTTPLVTNSGGATPTSIATPTSTPTTPVTPGLVSPPPELVDPNQHPLGLPLSRRQKSFNKCKTPLYNMEHTDELSIIGHNDSLEIQAVVSSGALYIHRCCLEFSPPFQATSSEEDLEQAEETRIRGIVTSALTRKCAFCTRHGASIPCKMSCNKYYHLPCLLASGGFMDFQSKGSFCKDHLYQVPLVCTSEIDCRTCRTIGDIANLMTCVVCGAHYHGTCVGLAQLPGVRSGWACRGCRVCQVCREPAPGEARAVCCDHCDKLYHAACLRPLMATVPKYGWKCKCCRVCSDCGSRSPGAGPSSRWHAHYTVCDSCYQQRNKGSCCPLCRRAYRAAAYRDMIRCTLCKRYVHGTCDPDAEPQQYRKNKDQNPSYEYSCPICKSQLALTGSKPGSFEEDSTASVSQDSSYGDDNSNLQDQDPLAIETKPDVGLGKGKPFSVSSKVAKKKIGGYKLKGGFPAGGKLGFQKRQRSLLDFGRKRASKPKMRGVFGVPGLGLQRPQAPDSKSSEDDPGMENKLVLCSSKDKFVLTQDLCVMCGAVGTDSEGCLIACAQCGQTYHPYCVNIKVIQN
Summary
Catalytic Activity
L-lysyl-[histone] + S-adenosyl-L-methionine = H(+) + N(6)-methyl-L-lysyl-[histone] + S-adenosyl-L-homocysteine
Uniprot
H9IZ17
A0A3S2NCM7
A0A212F1E6
A0A194QBS6
A0A2H1W9I8
A0A2A4IVZ3
+ More
A0A1B0CJ96 A0A182GXI3 A0A1B0DCK0 Q17A65 A0A182TGZ4 A0A1S4FAE3 A0A182ILT4 A0A182HFW5 A0A182JNE9 A0A084WCC3 A0A182NZI8 A0A182VKR8 A0A182X5J0 A0A182YEC8 B0WUW9 A0A1J1I3M1 A0A182QZL4 A0A336MLN0 A0A336MJT3 A0A336ML50 A0A336MIG1 A0A336MH66 A0A2A3E2H1 A0A1Y1L2Y7 A0A1Y1L3C7 A0A1Y1L8Z9 A0A1Y1L6R4 A0A087ZQZ5 A0A2M4A4V8 A0A2M4A549 A0A026WTW3 A0A139WMS0 D6W9D4 A0A182MGK2 A0A182FVU2 K7J2B3 A0A182WDF0 A0A0C9R071 A0A182RTG3 A0A182MXJ6 E0VS51 U4UBC4 A0A151IQH0 A0A195FJK3 F4WR72 A0A195BAP5 A0A1B6F6S2 A0A1B6GJ31 A0A1B6D156 N6TZM2 A0A1B6K3T0 A0A182LHR2 A0A2H8TLB4 J9JLG9 A0A2S2Q7Q2 A0A2S2QNC8 A0A023F0J6 E9IRP1 A0A0L0C8T6 A0A1I8MA62 A0A1I8MA66 A0A1B0A978 A0A1A9WP71 A0A1A9UJ12 A0A1B0FJW5 A0A151JCE8 A0A1I8NS86 T1GGQ9 B4KMC6 A0A0A1WV58 B4J645 A0A0K8UTG6 A0A0M4EBI1 W8C427 A0A0K8UPC3 A0A0K8V2M0 A0A0K8V3I4 B4LNY9 B3MCM2 T1IZ38
A0A1B0CJ96 A0A182GXI3 A0A1B0DCK0 Q17A65 A0A182TGZ4 A0A1S4FAE3 A0A182ILT4 A0A182HFW5 A0A182JNE9 A0A084WCC3 A0A182NZI8 A0A182VKR8 A0A182X5J0 A0A182YEC8 B0WUW9 A0A1J1I3M1 A0A182QZL4 A0A336MLN0 A0A336MJT3 A0A336ML50 A0A336MIG1 A0A336MH66 A0A2A3E2H1 A0A1Y1L2Y7 A0A1Y1L3C7 A0A1Y1L8Z9 A0A1Y1L6R4 A0A087ZQZ5 A0A2M4A4V8 A0A2M4A549 A0A026WTW3 A0A139WMS0 D6W9D4 A0A182MGK2 A0A182FVU2 K7J2B3 A0A182WDF0 A0A0C9R071 A0A182RTG3 A0A182MXJ6 E0VS51 U4UBC4 A0A151IQH0 A0A195FJK3 F4WR72 A0A195BAP5 A0A1B6F6S2 A0A1B6GJ31 A0A1B6D156 N6TZM2 A0A1B6K3T0 A0A182LHR2 A0A2H8TLB4 J9JLG9 A0A2S2Q7Q2 A0A2S2QNC8 A0A023F0J6 E9IRP1 A0A0L0C8T6 A0A1I8MA62 A0A1I8MA66 A0A1B0A978 A0A1A9WP71 A0A1A9UJ12 A0A1B0FJW5 A0A151JCE8 A0A1I8NS86 T1GGQ9 B4KMC6 A0A0A1WV58 B4J645 A0A0K8UTG6 A0A0M4EBI1 W8C427 A0A0K8UPC3 A0A0K8V2M0 A0A0K8V3I4 B4LNY9 B3MCM2 T1IZ38
EC Number
2.1.1.43
Pubmed
EMBL
BABH01010903
BABH01010904
BABH01010905
BABH01010906
BABH01010907
BABH01010908
+ More
RSAL01000222 RVE44033.1 AGBW02010898 OWR47544.1 KQ459220 KPJ02917.1 ODYU01007184 SOQ49738.1 NWSH01005542 PCG64137.1 AJWK01014351 AJWK01014352 JXUM01095568 KQ564203 KXJ72592.1 AJVK01005146 AJVK01005147 AJVK01005148 AJVK01005149 CH477339 EAT43161.1 AXCP01008506 APCN01005430 ATLV01022651 KE525335 KFB47867.1 DS232111 EDS35112.1 CVRI01000040 CRK94795.1 AXCN02001722 UFQT01001259 SSX29733.1 SSX29731.1 SSX29729.1 SSX29730.1 SSX29732.1 KZ288474 PBC25301.1 GEZM01066211 GEZM01066209 GEZM01066202 GEZM01066200 GEZM01066199 GEZM01066198 GEZM01066193 JAV68063.1 GEZM01066212 GEZM01066197 GEZM01066195 GEZM01066192 GEZM01066191 JAV68084.1 GEZM01066214 GEZM01066210 GEZM01066208 GEZM01066207 GEZM01066206 GEZM01066201 GEZM01066194 JAV68056.1 GEZM01066213 GEZM01066205 GEZM01066204 GEZM01066203 GEZM01066196 JAV68070.1 GGFK01002516 MBW35837.1 GGFK01002521 MBW35842.1 KK107119 EZA58544.1 KQ971312 KYB29184.1 EEZ98177.2 AXCM01003735 AAZX01001561 GBYB01009504 JAG79271.1 DS235745 EEB16207.1 KB632184 ERL89633.1 KQ976780 KYN08397.1 KQ981512 KYN40840.1 GL888284 EGI63299.1 KQ976532 KYM81621.1 GECZ01024206 JAS45563.1 GECZ01007317 JAS62452.1 GEDC01017945 JAS19353.1 APGK01048496 APGK01048497 KB741112 ENN73816.1 GECU01001608 JAT06099.1 GFXV01003024 MBW14829.1 ABLF02022838 ABLF02022842 ABLF02022849 GGMS01004541 MBY73744.1 GGMS01010062 MBY79265.1 GBBI01004228 JAC14484.1 GL765280 EFZ16759.1 JRES01000737 KNC28853.1 CCAG010018850 KQ979048 KYN23222.1 CAQQ02111077 CAQQ02111078 CAQQ02111079 CH933808 EDW09814.2 GBXI01011892 JAD02400.1 CH916367 EDW01903.1 GDHF01022320 JAI29994.1 CP012524 ALC42621.1 GAMC01002517 JAC04039.1 GDHF01023765 JAI28549.1 GDHF01019142 JAI33172.1 GDHF01018827 JAI33487.1 CH940648 EDW61158.1 CH902619 EDV36256.1 AFFK01020382
RSAL01000222 RVE44033.1 AGBW02010898 OWR47544.1 KQ459220 KPJ02917.1 ODYU01007184 SOQ49738.1 NWSH01005542 PCG64137.1 AJWK01014351 AJWK01014352 JXUM01095568 KQ564203 KXJ72592.1 AJVK01005146 AJVK01005147 AJVK01005148 AJVK01005149 CH477339 EAT43161.1 AXCP01008506 APCN01005430 ATLV01022651 KE525335 KFB47867.1 DS232111 EDS35112.1 CVRI01000040 CRK94795.1 AXCN02001722 UFQT01001259 SSX29733.1 SSX29731.1 SSX29729.1 SSX29730.1 SSX29732.1 KZ288474 PBC25301.1 GEZM01066211 GEZM01066209 GEZM01066202 GEZM01066200 GEZM01066199 GEZM01066198 GEZM01066193 JAV68063.1 GEZM01066212 GEZM01066197 GEZM01066195 GEZM01066192 GEZM01066191 JAV68084.1 GEZM01066214 GEZM01066210 GEZM01066208 GEZM01066207 GEZM01066206 GEZM01066201 GEZM01066194 JAV68056.1 GEZM01066213 GEZM01066205 GEZM01066204 GEZM01066203 GEZM01066196 JAV68070.1 GGFK01002516 MBW35837.1 GGFK01002521 MBW35842.1 KK107119 EZA58544.1 KQ971312 KYB29184.1 EEZ98177.2 AXCM01003735 AAZX01001561 GBYB01009504 JAG79271.1 DS235745 EEB16207.1 KB632184 ERL89633.1 KQ976780 KYN08397.1 KQ981512 KYN40840.1 GL888284 EGI63299.1 KQ976532 KYM81621.1 GECZ01024206 JAS45563.1 GECZ01007317 JAS62452.1 GEDC01017945 JAS19353.1 APGK01048496 APGK01048497 KB741112 ENN73816.1 GECU01001608 JAT06099.1 GFXV01003024 MBW14829.1 ABLF02022838 ABLF02022842 ABLF02022849 GGMS01004541 MBY73744.1 GGMS01010062 MBY79265.1 GBBI01004228 JAC14484.1 GL765280 EFZ16759.1 JRES01000737 KNC28853.1 CCAG010018850 KQ979048 KYN23222.1 CAQQ02111077 CAQQ02111078 CAQQ02111079 CH933808 EDW09814.2 GBXI01011892 JAD02400.1 CH916367 EDW01903.1 GDHF01022320 JAI29994.1 CP012524 ALC42621.1 GAMC01002517 JAC04039.1 GDHF01023765 JAI28549.1 GDHF01019142 JAI33172.1 GDHF01018827 JAI33487.1 CH940648 EDW61158.1 CH902619 EDV36256.1 AFFK01020382
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000218220
UP000092461
+ More
UP000069940 UP000249989 UP000092462 UP000008820 UP000075902 UP000075880 UP000075840 UP000075881 UP000030765 UP000075885 UP000075903 UP000076407 UP000076408 UP000002320 UP000183832 UP000075886 UP000242457 UP000005203 UP000053097 UP000007266 UP000075883 UP000069272 UP000002358 UP000075920 UP000075900 UP000075884 UP000009046 UP000030742 UP000078542 UP000078541 UP000007755 UP000078540 UP000019118 UP000075882 UP000007819 UP000037069 UP000095301 UP000092445 UP000091820 UP000078200 UP000092444 UP000078492 UP000095300 UP000015102 UP000009192 UP000001070 UP000092553 UP000008792 UP000007801
UP000069940 UP000249989 UP000092462 UP000008820 UP000075902 UP000075880 UP000075840 UP000075881 UP000030765 UP000075885 UP000075903 UP000076407 UP000076408 UP000002320 UP000183832 UP000075886 UP000242457 UP000005203 UP000053097 UP000007266 UP000075883 UP000069272 UP000002358 UP000075920 UP000075900 UP000075884 UP000009046 UP000030742 UP000078542 UP000078541 UP000007755 UP000078540 UP000019118 UP000075882 UP000007819 UP000037069 UP000095301 UP000092445 UP000091820 UP000078200 UP000092444 UP000078492 UP000095300 UP000015102 UP000009192 UP000001070 UP000092553 UP000008792 UP000007801
PRIDE
Pfam
Interpro
IPR003888
FYrich_N
+ More
IPR034732 EPHD
IPR036910 HMG_box_dom_sf
IPR001841 Znf_RING
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001214 SET_dom
IPR009071 HMG_box_dom
IPR003889 FYrich_C
IPR003616 Post-SET_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR000637 HMGI/Y_DNA-bd_CS
IPR019786 Zinc_finger_PHD-type_CS
IPR000679 Znf_GATA
IPR037877 KMT2C
IPR009286 Ins_P5_2-kin
IPR034732 EPHD
IPR036910 HMG_box_dom_sf
IPR001841 Znf_RING
IPR011011 Znf_FYVE_PHD
IPR019787 Znf_PHD-finger
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001214 SET_dom
IPR009071 HMG_box_dom
IPR003889 FYrich_C
IPR003616 Post-SET_dom
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR000637 HMGI/Y_DNA-bd_CS
IPR019786 Zinc_finger_PHD-type_CS
IPR000679 Znf_GATA
IPR037877 KMT2C
IPR009286 Ins_P5_2-kin
Gene 3D
ProteinModelPortal
H9IZ17
A0A3S2NCM7
A0A212F1E6
A0A194QBS6
A0A2H1W9I8
A0A2A4IVZ3
+ More
A0A1B0CJ96 A0A182GXI3 A0A1B0DCK0 Q17A65 A0A182TGZ4 A0A1S4FAE3 A0A182ILT4 A0A182HFW5 A0A182JNE9 A0A084WCC3 A0A182NZI8 A0A182VKR8 A0A182X5J0 A0A182YEC8 B0WUW9 A0A1J1I3M1 A0A182QZL4 A0A336MLN0 A0A336MJT3 A0A336ML50 A0A336MIG1 A0A336MH66 A0A2A3E2H1 A0A1Y1L2Y7 A0A1Y1L3C7 A0A1Y1L8Z9 A0A1Y1L6R4 A0A087ZQZ5 A0A2M4A4V8 A0A2M4A549 A0A026WTW3 A0A139WMS0 D6W9D4 A0A182MGK2 A0A182FVU2 K7J2B3 A0A182WDF0 A0A0C9R071 A0A182RTG3 A0A182MXJ6 E0VS51 U4UBC4 A0A151IQH0 A0A195FJK3 F4WR72 A0A195BAP5 A0A1B6F6S2 A0A1B6GJ31 A0A1B6D156 N6TZM2 A0A1B6K3T0 A0A182LHR2 A0A2H8TLB4 J9JLG9 A0A2S2Q7Q2 A0A2S2QNC8 A0A023F0J6 E9IRP1 A0A0L0C8T6 A0A1I8MA62 A0A1I8MA66 A0A1B0A978 A0A1A9WP71 A0A1A9UJ12 A0A1B0FJW5 A0A151JCE8 A0A1I8NS86 T1GGQ9 B4KMC6 A0A0A1WV58 B4J645 A0A0K8UTG6 A0A0M4EBI1 W8C427 A0A0K8UPC3 A0A0K8V2M0 A0A0K8V3I4 B4LNY9 B3MCM2 T1IZ38
A0A1B0CJ96 A0A182GXI3 A0A1B0DCK0 Q17A65 A0A182TGZ4 A0A1S4FAE3 A0A182ILT4 A0A182HFW5 A0A182JNE9 A0A084WCC3 A0A182NZI8 A0A182VKR8 A0A182X5J0 A0A182YEC8 B0WUW9 A0A1J1I3M1 A0A182QZL4 A0A336MLN0 A0A336MJT3 A0A336ML50 A0A336MIG1 A0A336MH66 A0A2A3E2H1 A0A1Y1L2Y7 A0A1Y1L3C7 A0A1Y1L8Z9 A0A1Y1L6R4 A0A087ZQZ5 A0A2M4A4V8 A0A2M4A549 A0A026WTW3 A0A139WMS0 D6W9D4 A0A182MGK2 A0A182FVU2 K7J2B3 A0A182WDF0 A0A0C9R071 A0A182RTG3 A0A182MXJ6 E0VS51 U4UBC4 A0A151IQH0 A0A195FJK3 F4WR72 A0A195BAP5 A0A1B6F6S2 A0A1B6GJ31 A0A1B6D156 N6TZM2 A0A1B6K3T0 A0A182LHR2 A0A2H8TLB4 J9JLG9 A0A2S2Q7Q2 A0A2S2QNC8 A0A023F0J6 E9IRP1 A0A0L0C8T6 A0A1I8MA62 A0A1I8MA66 A0A1B0A978 A0A1A9WP71 A0A1A9UJ12 A0A1B0FJW5 A0A151JCE8 A0A1I8NS86 T1GGQ9 B4KMC6 A0A0A1WV58 B4J645 A0A0K8UTG6 A0A0M4EBI1 W8C427 A0A0K8UPC3 A0A0K8V2M0 A0A0K8V3I4 B4LNY9 B3MCM2 T1IZ38
PDB
2YSM
E-value=2.13652e-19,
Score=238
Ontologies
GO
GO:0042800
GO:0046872
GO:0035097
GO:0018024
GO:0005634
GO:0044666
GO:0003677
GO:0003713
GO:0045944
GO:0042393
GO:0003676
GO:0006355
GO:0016021
GO:0003682
GO:0035076
GO:0005700
GO:0018990
GO:1900114
GO:0030374
GO:0008230
GO:0007474
GO:0000790
GO:0008586
GO:0043565
GO:0008270
GO:0005524
GO:0035299
GO:0005515
GO:0045454
GO:0051920
GO:0003887
GO:0001664
GO:0005525
GO:0016491
GO:0003824
PANTHER
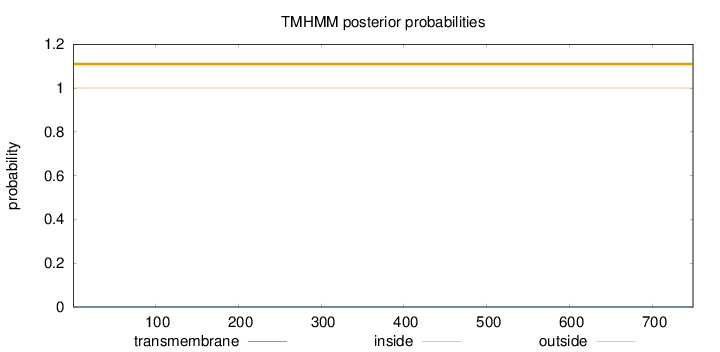
Topology
Subcellular location
Nucleus
Length:
749
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00238
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00008
outside
1 - 749
Population Genetic Test Statistics
Pi
163.509393
Theta
158.905019
Tajima's D
-0.806418
CLR
159.611833
CSRT
0.176441177941103
Interpretation
Uncertain
