Gene
KWMTBOMO05274
Pre Gene Modal
BGIBMGA002407
Annotation
hypothetical_protein_KGM_14799_[Danaus_plexippus]
Full name
Sulfurtransferase
Location in the cell
Nuclear Reliability : 2.919
Sequence
CDS
ATGGTTGATCCGCGACGAGTAGTAAGCTACGAAGATGTATTGAAAGTTATTCATCAACCCGAAAAGGTGTTGATCGACGTGCGTAATCCTGATGAAGTAAACACAACTGGAAAAATACCTTCAAGCATCAATATTCCACTAAATAGTGTTCAAGAGGTTCTGGTATCAATGTCTAACGAGGAGTTCAAGAAACAATATCAGAGAAACAAACCTAGTTCAACAGACGAACTGATATTTTACTGTCAATCTGGAAGAAGATCAACTGAGGCATTGGACATAGCATTAAAGTTAGGTTATTCCAAGTCGAAAACATATTTAGGGAGCTGGAATGAATGGTCTAGAACACAAAAATAA
Protein
MVDPRRVVSYEDVLKVIHQPEKVLIDVRNPDEVNTTGKIPSSINIPLNSVQEVLVSMSNEEFKKQYQRNKPSSTDELIFYCQSGRRSTEALDIALKLGYSKSKTYLGSWNEWSRTQK
Summary
Feature
chain Sulfurtransferase
Uniprot
H9IYS2
A0A3S2LU62
S4P4E5
I4DKS8
A0A345BJE5
A0A212F1D9
+ More
A0A2H1W9K2 A0A0L7KSW0 A0A1L8DUC5 W5JD98 F5HIX9 A0A2M4C498 A0A2M4AXY3 A0A182LV89 A0A2M3ZBF6 A0A2M3ZB53 A0A1A9VR23 A0A182XZ05 H9IZ16 A0A182WJ42 A0A084WNG1 Q178I4 Q178W5 A0A1X9JSU3 A0A1I8NC47 T1PDV6 A0A1S4FBL6 A0A1A9ZLA4 A0A1B0G855 A0A1S4FBV1 A0A1I8NDC6 A0A1L8EDH8 A0A182JMS2 A0A023EGL2 B0X3A2 A0A1Q3FGA6 A0A2M4AYG2 A0A1L8ED68 A0A1L8EDG3 A0A2M3ZIY2 E0VS08 B0X3A3 U5ETS5 B0X3A1 A0A1J1IYI5 A0A1I8P6X1 A0A1L8DU43 A0A1A9ZLA2 A0A336MVY0 A0A336LK15 B4NBN9 A0A0L0CJ64 A0A3B0K9I3 A0A1A9VR25 A0A1I8PHC2 A0A1A9W5T8 A0A139WI34 D6WIZ1 A0A336MY58 A0A2H1W9I4 A0A1Q3F8G3 A0A1Q3G437 A0A0T6BHK1 B3M2S5 A0A212F1E1 B4LGT3 A0A345BJE4 A0A0T6AZC3 A0A139WAY7 A0A1A9XDK2 A0A1A9XI35 A0A1A9VS48 A0A0L7KST6 A0A1B0BJP6 A0A1B0BJP8 A0A1A9XDL3 A0A1B0BTV6 A0A1A9ZGD5 A0A1A9W5T9 A7URA9 B4MN42 G1PYL7 B5DVK7 A0A0T6B0V8 A7URA7 B4L116 A0A0L0CQ49 B4MBZ8 A0A0N1INN3 B4G432 A0A1I8MFJ7 A0A194QBX0 A0A1Y9TFG1 B4K5P3 A0A0T6B033 S7N1R8 A0A1Y2GUU7 A0A2M3ZC83 A0A182PHA4
A0A2H1W9K2 A0A0L7KSW0 A0A1L8DUC5 W5JD98 F5HIX9 A0A2M4C498 A0A2M4AXY3 A0A182LV89 A0A2M3ZBF6 A0A2M3ZB53 A0A1A9VR23 A0A182XZ05 H9IZ16 A0A182WJ42 A0A084WNG1 Q178I4 Q178W5 A0A1X9JSU3 A0A1I8NC47 T1PDV6 A0A1S4FBL6 A0A1A9ZLA4 A0A1B0G855 A0A1S4FBV1 A0A1I8NDC6 A0A1L8EDH8 A0A182JMS2 A0A023EGL2 B0X3A2 A0A1Q3FGA6 A0A2M4AYG2 A0A1L8ED68 A0A1L8EDG3 A0A2M3ZIY2 E0VS08 B0X3A3 U5ETS5 B0X3A1 A0A1J1IYI5 A0A1I8P6X1 A0A1L8DU43 A0A1A9ZLA2 A0A336MVY0 A0A336LK15 B4NBN9 A0A0L0CJ64 A0A3B0K9I3 A0A1A9VR25 A0A1I8PHC2 A0A1A9W5T8 A0A139WI34 D6WIZ1 A0A336MY58 A0A2H1W9I4 A0A1Q3F8G3 A0A1Q3G437 A0A0T6BHK1 B3M2S5 A0A212F1E1 B4LGT3 A0A345BJE4 A0A0T6AZC3 A0A139WAY7 A0A1A9XDK2 A0A1A9XI35 A0A1A9VS48 A0A0L7KST6 A0A1B0BJP6 A0A1B0BJP8 A0A1A9XDL3 A0A1B0BTV6 A0A1A9ZGD5 A0A1A9W5T9 A7URA9 B4MN42 G1PYL7 B5DVK7 A0A0T6B0V8 A7URA7 B4L116 A0A0L0CQ49 B4MBZ8 A0A0N1INN3 B4G432 A0A1I8MFJ7 A0A194QBX0 A0A1Y9TFG1 B4K5P3 A0A0T6B033 S7N1R8 A0A1Y2GUU7 A0A2M3ZC83 A0A182PHA4
Pubmed
EMBL
BABH01010899
RSAL01000222
RVE44029.1
GAIX01006004
JAA86556.1
AK401896
+ More
KQ459220 BAM18518.1 KPJ02915.1 MH036752 AXF50551.1 AGBW02010898 OWR47546.1 ODYU01007184 SOQ49740.1 JTDY01006302 KOB66124.1 GFDF01004078 JAV10006.1 ADMH02001493 ETN62322.1 AAAB01008807 EGK96240.1 GGFJ01010830 MBW59971.1 GGFK01012284 MBW45605.1 AXCM01001159 GGFM01005037 MBW25788.1 GGFM01005023 MBW25774.1 BABH01010895 ATLV01024599 KE525352 KFB51755.1 CH477363 EAT42598.1 CH477358 EAT42734.1 KX344893 AQY54360.1 KA646118 AFP60747.1 CCAG010000199 GFDG01002095 JAV16704.1 GAPW01005654 JAC07944.1 DS232311 EDS39604.1 GFDL01008513 JAV26532.1 GGFK01012431 MBW45752.1 GFDG01002122 JAV16677.1 GFDG01002103 JAV16696.1 GGFM01007740 MBW28491.1 DS235677 EEB16164.1 EDS39605.1 GANO01001772 JAB58099.1 EDS39603.1 CVRI01000062 CRL04206.1 GFDF01004112 JAV09972.1 UFQT01003095 SSX34552.1 UFQT01000033 SSX18286.1 CH964232 EDW81203.1 JRES01000321 KNC32295.1 OUUW01000007 SPP82729.1 KQ971342 KYB27559.1 EFA03724.2 SSX34551.1 SOQ49741.1 GFDL01011226 JAV23819.1 GFDL01000484 JAV34561.1 LJIG01000152 KRT86788.1 CH902617 EDV42396.2 OWR47547.1 CH940647 EDW70548.2 MH036751 AXF50550.1 LJIG01022459 KRT80470.1 KQ971379 KYB25070.1 KOB66126.1 JXJN01015477 JXJN01015478 JXJN01020372 EDO64551.1 CH963847 EDW73598.2 AAPE02031043 CM000070 EDY68043.1 KRT80471.1 EDO64549.1 EDO64550.1 CH933809 EDW18173.2 JRES01000062 KNC34490.1 CH940656 EDW58619.1 KQ461135 KPJ08854.1 CH479179 EDW24443.1 KPJ02914.1 KX344892 AQY54359.1 CH933806 EDW15105.1 KRT80469.1 KE163131 EPQ10939.1 MCFF01000010 ORZ22742.1 GGFM01005416 MBW26167.1
KQ459220 BAM18518.1 KPJ02915.1 MH036752 AXF50551.1 AGBW02010898 OWR47546.1 ODYU01007184 SOQ49740.1 JTDY01006302 KOB66124.1 GFDF01004078 JAV10006.1 ADMH02001493 ETN62322.1 AAAB01008807 EGK96240.1 GGFJ01010830 MBW59971.1 GGFK01012284 MBW45605.1 AXCM01001159 GGFM01005037 MBW25788.1 GGFM01005023 MBW25774.1 BABH01010895 ATLV01024599 KE525352 KFB51755.1 CH477363 EAT42598.1 CH477358 EAT42734.1 KX344893 AQY54360.1 KA646118 AFP60747.1 CCAG010000199 GFDG01002095 JAV16704.1 GAPW01005654 JAC07944.1 DS232311 EDS39604.1 GFDL01008513 JAV26532.1 GGFK01012431 MBW45752.1 GFDG01002122 JAV16677.1 GFDG01002103 JAV16696.1 GGFM01007740 MBW28491.1 DS235677 EEB16164.1 EDS39605.1 GANO01001772 JAB58099.1 EDS39603.1 CVRI01000062 CRL04206.1 GFDF01004112 JAV09972.1 UFQT01003095 SSX34552.1 UFQT01000033 SSX18286.1 CH964232 EDW81203.1 JRES01000321 KNC32295.1 OUUW01000007 SPP82729.1 KQ971342 KYB27559.1 EFA03724.2 SSX34551.1 SOQ49741.1 GFDL01011226 JAV23819.1 GFDL01000484 JAV34561.1 LJIG01000152 KRT86788.1 CH902617 EDV42396.2 OWR47547.1 CH940647 EDW70548.2 MH036751 AXF50550.1 LJIG01022459 KRT80470.1 KQ971379 KYB25070.1 KOB66126.1 JXJN01015477 JXJN01015478 JXJN01020372 EDO64551.1 CH963847 EDW73598.2 AAPE02031043 CM000070 EDY68043.1 KRT80471.1 EDO64549.1 EDO64550.1 CH933809 EDW18173.2 JRES01000062 KNC34490.1 CH940656 EDW58619.1 KQ461135 KPJ08854.1 CH479179 EDW24443.1 KPJ02914.1 KX344892 AQY54359.1 CH933806 EDW15105.1 KRT80469.1 KE163131 EPQ10939.1 MCFF01000010 ORZ22742.1 GGFM01005416 MBW26167.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000037510
UP000000673
+ More
UP000007062 UP000075883 UP000078200 UP000076408 UP000075920 UP000030765 UP000008820 UP000095301 UP000092445 UP000092444 UP000075880 UP000002320 UP000009046 UP000183832 UP000095300 UP000007798 UP000037069 UP000268350 UP000091820 UP000007266 UP000007801 UP000008792 UP000092443 UP000092460 UP000001074 UP000001819 UP000009192 UP000053240 UP000008744 UP000193648 UP000075885
UP000007062 UP000075883 UP000078200 UP000076408 UP000075920 UP000030765 UP000008820 UP000095301 UP000092445 UP000092444 UP000075880 UP000002320 UP000009046 UP000183832 UP000095300 UP000007798 UP000037069 UP000268350 UP000091820 UP000007266 UP000007801 UP000008792 UP000092443 UP000092460 UP000001074 UP000001819 UP000009192 UP000053240 UP000008744 UP000193648 UP000075885
Pfam
PF00581 Rhodanese
Interpro
SUPFAM
SSF52821
SSF52821
Gene 3D
ProteinModelPortal
H9IYS2
A0A3S2LU62
S4P4E5
I4DKS8
A0A345BJE5
A0A212F1D9
+ More
A0A2H1W9K2 A0A0L7KSW0 A0A1L8DUC5 W5JD98 F5HIX9 A0A2M4C498 A0A2M4AXY3 A0A182LV89 A0A2M3ZBF6 A0A2M3ZB53 A0A1A9VR23 A0A182XZ05 H9IZ16 A0A182WJ42 A0A084WNG1 Q178I4 Q178W5 A0A1X9JSU3 A0A1I8NC47 T1PDV6 A0A1S4FBL6 A0A1A9ZLA4 A0A1B0G855 A0A1S4FBV1 A0A1I8NDC6 A0A1L8EDH8 A0A182JMS2 A0A023EGL2 B0X3A2 A0A1Q3FGA6 A0A2M4AYG2 A0A1L8ED68 A0A1L8EDG3 A0A2M3ZIY2 E0VS08 B0X3A3 U5ETS5 B0X3A1 A0A1J1IYI5 A0A1I8P6X1 A0A1L8DU43 A0A1A9ZLA2 A0A336MVY0 A0A336LK15 B4NBN9 A0A0L0CJ64 A0A3B0K9I3 A0A1A9VR25 A0A1I8PHC2 A0A1A9W5T8 A0A139WI34 D6WIZ1 A0A336MY58 A0A2H1W9I4 A0A1Q3F8G3 A0A1Q3G437 A0A0T6BHK1 B3M2S5 A0A212F1E1 B4LGT3 A0A345BJE4 A0A0T6AZC3 A0A139WAY7 A0A1A9XDK2 A0A1A9XI35 A0A1A9VS48 A0A0L7KST6 A0A1B0BJP6 A0A1B0BJP8 A0A1A9XDL3 A0A1B0BTV6 A0A1A9ZGD5 A0A1A9W5T9 A7URA9 B4MN42 G1PYL7 B5DVK7 A0A0T6B0V8 A7URA7 B4L116 A0A0L0CQ49 B4MBZ8 A0A0N1INN3 B4G432 A0A1I8MFJ7 A0A194QBX0 A0A1Y9TFG1 B4K5P3 A0A0T6B033 S7N1R8 A0A1Y2GUU7 A0A2M3ZC83 A0A182PHA4
A0A2H1W9K2 A0A0L7KSW0 A0A1L8DUC5 W5JD98 F5HIX9 A0A2M4C498 A0A2M4AXY3 A0A182LV89 A0A2M3ZBF6 A0A2M3ZB53 A0A1A9VR23 A0A182XZ05 H9IZ16 A0A182WJ42 A0A084WNG1 Q178I4 Q178W5 A0A1X9JSU3 A0A1I8NC47 T1PDV6 A0A1S4FBL6 A0A1A9ZLA4 A0A1B0G855 A0A1S4FBV1 A0A1I8NDC6 A0A1L8EDH8 A0A182JMS2 A0A023EGL2 B0X3A2 A0A1Q3FGA6 A0A2M4AYG2 A0A1L8ED68 A0A1L8EDG3 A0A2M3ZIY2 E0VS08 B0X3A3 U5ETS5 B0X3A1 A0A1J1IYI5 A0A1I8P6X1 A0A1L8DU43 A0A1A9ZLA2 A0A336MVY0 A0A336LK15 B4NBN9 A0A0L0CJ64 A0A3B0K9I3 A0A1A9VR25 A0A1I8PHC2 A0A1A9W5T8 A0A139WI34 D6WIZ1 A0A336MY58 A0A2H1W9I4 A0A1Q3F8G3 A0A1Q3G437 A0A0T6BHK1 B3M2S5 A0A212F1E1 B4LGT3 A0A345BJE4 A0A0T6AZC3 A0A139WAY7 A0A1A9XDK2 A0A1A9XI35 A0A1A9VS48 A0A0L7KST6 A0A1B0BJP6 A0A1B0BJP8 A0A1A9XDL3 A0A1B0BTV6 A0A1A9ZGD5 A0A1A9W5T9 A7URA9 B4MN42 G1PYL7 B5DVK7 A0A0T6B0V8 A7URA7 B4L116 A0A0L0CQ49 B4MBZ8 A0A0N1INN3 B4G432 A0A1I8MFJ7 A0A194QBX0 A0A1Y9TFG1 B4K5P3 A0A0T6B033 S7N1R8 A0A1Y2GUU7 A0A2M3ZC83 A0A182PHA4
PDB
3D1P
E-value=4.64551e-16,
Score=199
Ontologies
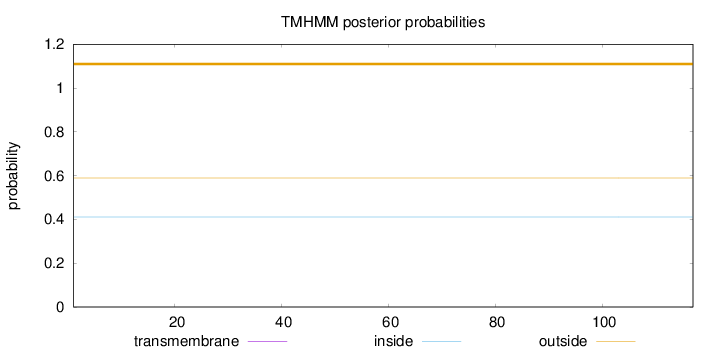
Topology
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.41060
outside
1 - 117
Population Genetic Test Statistics
Pi
458.392123
Theta
193.204072
Tajima's D
4.328279
CLR
0.5425
CSRT
0.99970001499925
Interpretation
Uncertain
