Pre Gene Modal
BGIBMGA002413
Annotation
Contactin_[Operophtera_brumata]
Full name
Succinate--CoA ligase [GDP-forming] subunit beta, mitochondrial
+ More
Coronin
Contactin
Coronin
Contactin
Alternative Name
GTP-specific succinyl-CoA synthetase subunit beta
Succinyl-CoA synthetase beta-G chain
Succinyl-CoA synthetase beta-G chain
Location in the cell
Extracellular Reliability : 1.119
Sequence
CDS
ATGAAGTGTACGTTCCTTGTACTCGCAGTTGCGAGTTTGGTGAGGACCCAAGCACCTGAAAATGGTATCTATAATCAGAACTATCCGGAAGGAAACCAGAATAATACACAATATCGTCCCGGGCCCCAAATCCCTTACGGCAGTTTTCCCGTAAATAATGAATATCAACAGCCACCTCAAAGCTCTTTCTACCAGGACAGCCTCTACAACAGTGACAGCTATGCTGAAAATGAAAATGATGTCGAAAATACTGCGAGTGTATATGCTGGCTTAGATTACGAAGAGAAGTACCGATGTCCACCCAACTGGCTCCGCTACAAAGAGTCATGTTATCGGTTTACGAAGTCTCCTGATCGTCCTAGAAATGAAGCAAGAAAAATATGCCAAGCCTATGAAGCTGATTTAGTAGCCGTCAATAGCCCTGAAGAACATGGTTTTATAATAAACAATTTAATGAGGCTCGACCCTCAGAAGGGTTCATGGTATATAGGAGCTCACCAACAGTCTCCAGGTTATTGGTCCAATGATTATGATGGTTCACAGTTGACAGGTCTTGAGAATGCTTTCTTCGACGATCGACAATTTTCATTAAATAGTTATAATATATTTCCTAAAGATTTTTTGGTATACAAATTTTCACATCAAGATGGACGCTGGGGCTTAGCTGCTGTAGAAGGTACACAATATTATCACTTTATATGTGAGGGGCAAATGCAGCGTTTACATTATTTAATAGAAGAAGAAAGATCATTCACTTATGGCTTAGATACTTATGATCCAGAAAGGATTCCGAGAGGTCCATATTTTGTAAAACAGCCGGAAGATACTGTTTATGATCCTTTAAGAAATCATTATGTCCAACTTACTTGTATAGCAGGAGGATTTCCAGCTCCAACTTACAAATGGTTCGGAGAAGATTATCAAGTTGACAGTCCAGTTTACACAGAAATAGATCCTTTAGCTGACAGTAGGATTATATTAAGTGGAGGAACTTTAATGATTTATAACCCTGACAGTGAAAAGGATAATACAAAATATCACTGTACAGCTACAAACAAGTTTGGTACGATTAGATCAGAATCTGTCCGGTTGTCATTTGGATTCATAAGAGATTTCAATCAAAAGAGGCCAGTAGAAAGTGGAAATCAATACTGGGGTAAAGTAATGTATTGTGATCCTCCTTCGTACTATCCAGCTGTAAATATTCTGTGGGCTCGGAATTATTTCCCTAACCTTGTGGAAGAAGATAAACGTGTCTTTGTGTCATATGACGGTGGCCTCTATTTCTCAGCTTTAGAAACAATAGACCGAGGCAACTATAGTTGTAATGTAATGAGTAAAGTGTCTGATGAAGGCAGAAATGGTCCATTCTTTCCCTTACAGGTTTATCCGCATTCAGATTATCAGCAATTGAAATTCCCAAATAGCTTTCCTAAAGTATTTCCTGAAGCTCCAGTGGTTGGGCAAGAAGTCCGCCTGGAATGTGTCACATTTGGCTACCCAGTACCTTCTTATAATTGGACCAGACAGAATGGCCTTATACCTAAAAAAGCTCAATTGAGTAATTTTGACCGGGTTTTAACAATACCCAATGTAGGTGTAGAGGATGAAGGCGAATATATTTGTGATGTTCACAATGACAGAGCGAGGATATCAAATAGTGTGTATTTAAAAGTACAAGCTGAACCAAATTTTACAATTCCTCTTGGCGATAAACATATGGACAACAAAGGAGAATTACATTGGAACTGTGAGGCATTTGGGATACCAGATGTTAATTACACATGGTGGAAAAATGGTAAAAAACTAAAAAAAGATAATCTAGAACCACAGGACCGTGATAGGTACATTATTCAAGACAATGTTTTGATTATAAAATATTTAGACCCAACAAGAGATCCTGGAATGTACCAGTGTGAAGCAAAGAATCAATTGAAAGCTAGTTACTCGTCAGCGCAATTACGAGTGCTCTCTTTGAAACCATCATTTAAAAAACATCCTTTAGAATTGGAGACATACGGTTCTGAAGGTGGAAATGTGACATTAAAATGCAAACCAGAAGCTGCCCCAAAGCCAACATTCACTTGGAAAAAAGACAACATAGTTATTGGGGCAGGAGGAAAAAGATTCATTACCGAAAATGGGAATTTAATTATTAGACAACTTTCCAGAGATGATGAGGGCGTGTACACATGCGTCGCAAAGAATCAATACGGCACTGATGAGAGTAGAGGTCGTCTAATTGTCTTACGTGCACCACGTTTCCTTGAAAAACTCCCCCCTCGGATCACTGCACAAGTGAATTCAGTAGTTTTCTTGCACTGTAATGCCGATGTAGATCCTATTTTAGATACAGCGTATCTATGGAACCATAATGGCTTGCGGATTAAAGACTCATCAGATCTGTATGTCAACAAAAGAATAAAAATTGACGGTGGTGAGCTAACTATTTACAATGTAAGTCTTTCGGATGTGGGAGATTACGAATGTGTCGTTAAGTCCGCTATCGGAAGAATCTCTTCTCATGTGCAGCTGCGTATAGAGGGCCCGCCAGGTCCGCCGGGAGGCGTACAAATACTTGGCATACAGAGGTCTTCGGTGACCCTAGAGTGGACCGACAGTAACTCAAACGGTCGCCCTATAACTGGATACATGCTGACCGGACGCACCCACTGGAACTCAACTTGGTACGTGATCAGCGAGAATATACCCAACGTAATGGAAGTCGATAGATACAATGGGCGTAAACGAGCCACTGTTTCCACGACGCTTCTACCGTGGGCCATATATGAGTTTAGAATTCAAGCTATTAATATACTGGGTCCGGGGCAACCGTCCGCGCCGAGTCCGCAGTTCTCAACGTCGGGAGACAAGCCCTATGAGGCGCCCGCGAACGTTAGCGGCGGCGGCGGGAAGACCGGTGACCTCACAATCACGTGGACGCCCTTACCGACCTCCCTCCAAAACGGACCCGGCATACATTACAAAATATTCTGGCGCCGGAACGGCTCCGAAGTCGAATTTCAGTCCTTGCTTCTTAAGAAGTACGGAAACATCGGGACATACGTCGTGCACATATCTTCCACGTATTTCTACACGCCGTACGATGTCAAAGTACAAGCCTTCAACGATATAGGTCCCGGGCCAGAGAGCGAAGTAGTAACCATTTATTCGGCGGAGGACATGCCGCAGGTCGCGCCGCAGCTGGTCTCCGCGCGGTCCTTTAACTCCACAGCCCTGAACGTGACCTGGAACCCGATCGATCAGTCGCGCGAGAGGCTGCGCGGCAAGCTGATAGGCCACCGTCTCAAGTACTGGAAACAGGAGAACAAAGAAGAGGAGTGCATCTACTATTTGTCTCGAACGACTCGAAATTGGGCTCTTATAGTTGGTCTCCAGCCAGATACATATTATTATGTGAAGGTATAA
Protein
MKCTFLVLAVASLVRTQAPENGIYNQNYPEGNQNNTQYRPGPQIPYGSFPVNNEYQQPPQSSFYQDSLYNSDSYAENENDVENTASVYAGLDYEEKYRCPPNWLRYKESCYRFTKSPDRPRNEARKICQAYEADLVAVNSPEEHGFIINNLMRLDPQKGSWYIGAHQQSPGYWSNDYDGSQLTGLENAFFDDRQFSLNSYNIFPKDFLVYKFSHQDGRWGLAAVEGTQYYHFICEGQMQRLHYLIEEERSFTYGLDTYDPERIPRGPYFVKQPEDTVYDPLRNHYVQLTCIAGGFPAPTYKWFGEDYQVDSPVYTEIDPLADSRIILSGGTLMIYNPDSEKDNTKYHCTATNKFGTIRSESVRLSFGFIRDFNQKRPVESGNQYWGKVMYCDPPSYYPAVNILWARNYFPNLVEEDKRVFVSYDGGLYFSALETIDRGNYSCNVMSKVSDEGRNGPFFPLQVYPHSDYQQLKFPNSFPKVFPEAPVVGQEVRLECVTFGYPVPSYNWTRQNGLIPKKAQLSNFDRVLTIPNVGVEDEGEYICDVHNDRARISNSVYLKVQAEPNFTIPLGDKHMDNKGELHWNCEAFGIPDVNYTWWKNGKKLKKDNLEPQDRDRYIIQDNVLIIKYLDPTRDPGMYQCEAKNQLKASYSSAQLRVLSLKPSFKKHPLELETYGSEGGNVTLKCKPEAAPKPTFTWKKDNIVIGAGGKRFITENGNLIIRQLSRDDEGVYTCVAKNQYGTDESRGRLIVLRAPRFLEKLPPRITAQVNSVVFLHCNADVDPILDTAYLWNHNGLRIKDSSDLYVNKRIKIDGGELTIYNVSLSDVGDYECVVKSAIGRISSHVQLRIEGPPGPPGGVQILGIQRSSVTLEWTDSNSNGRPITGYMLTGRTHWNSTWYVISENIPNVMEVDRYNGRKRATVSTTLLPWAIYEFRIQAINILGPGQPSAPSPQFSTSGDKPYEAPANVSGGGGKTGDLTITWTPLPTSLQNGPGIHYKIFWRRNGSEVEFQSLLLKKYGNIGTYVVHISSTYFYTPYDVKVQAFNDIGPGPESEVVTIYSAEDMPQVAPQLVSARSFNSTALNVTWNPIDQSRERLRGKLIGHRLKYWKQENKEEECIYYLSRTTRNWALIVGLQPDTYYYVKV
Summary
Description
GTP-specific succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The beta subunit provides nucleotide specificity of the enzyme and binds the substrate succinate, while the binding sites for coenzyme A and phosphate are found in the alpha subunit.
Required for organization of septate junctions and paracellular barrier functions. Septate junctions, which are the equivalent of vertebrates tight junctions, are characterized by regular arrays of transverse structures that span the intermembrane space and form a physical barrier to diffusion.
Required for organization of septate junctions and paracellular barrier functions. Septate junctions, which are the equivalent of vertebrates tight junctions, are characterized by regular arrays of transverse structures that span the intermembrane space and form a physical barrier to diffusion.
Catalytic Activity
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
Cofactor
Mg(2+)
Subunit
Heterodimer of an alpha and a beta subunit. The beta subunit determines specificity for GTP.
Forms a complex with Nrg and Nrx.
Forms a complex with Nrg and Nrx.
Similarity
Belongs to the succinate/malate CoA ligase beta subunit family. GTP-specific subunit beta subfamily.
Belongs to the WD repeat coronin family.
Belongs to the immunoglobulin superfamily. Contactin family.
Belongs to the WD repeat coronin family.
Belongs to the immunoglobulin superfamily. Contactin family.
Keywords
Cell adhesion
Cell junction
Cell membrane
Complete proteome
Disulfide bond
Glycoprotein
GPI-anchor
Immunoglobulin domain
Lipoprotein
Membrane
Reference proteome
Repeat
Signal
Feature
chain Succinate--CoA ligase [GDP-forming] subunit beta, mitochondrial
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
A0A2H1VT68
A0A0L7LJS0
A0A2A4IXP8
B2DBK6
A0A194QBI6
A0A3S2LTD0
+ More
A0A1B3IIF8 A0A1B6ENC1 A0A2J7QMK8 A0A1B6CPI8 A0A067R1K6 A0A0A9W5Z2 A0A0K8SZ50 A0A1Y1ME68 A0A1Y1MCY1 A0A195EU65 A0A026WIX4 A0A195B0J4 A0A154PRQ2 A0A158NQW5 A0A195CM68 A0A195E5P9 F4WP72 A0A087ZZ67 A0A2A3EGS0 A0A232EZ17 A0A151WM91 E0VHE2 A0A224X5J2 N6T8T5 E2BBG0 A0A0C9QB16 A0A2S2Q4I6 A0A2H8TTG8 A0A2S2NZ57 X1XBF9 A0A139WNP4 A0A0M9A8L9 A0A139WNQ0 A0A310SDU4 W4VRM9 A0A3Q0IPR7 A0A3Q0JHC2 A0A336LRD9 A0A034VYW9 A0A0L0BR94 A0A1D2MXX3 W8B851 C4IXZ0 B4I3N0 A0A0A1WYZ4 A0A1I8NGE1 Q9VN14 A0A1W4VLC8 B4NID8 A0A1J1HHK2 A0A0P5RFZ7 A0A0N8E3P1 A0A1I8QBD3 A0A0P5MYZ4 B4K8Y9 A0A0N8BPK1 A0A0P6G4L4 A0A0P6H0T8 A0A0P6G5D7 A0A0P5S5M2 A0A0P5P8J5 A0A0P5QYP0 A0A0P5MLA3 A0A0P5JZS8 A0A0P5R4J0 A0A0P6D687 A0A0P4ZQ78 A0A1S4FFB1 A0A3B0KEU5 A0A0N8BQQ1 Q174B0 B4PTR6 A0A3B0JRG3 A0A0P4YSM5 B3LY10 A0A1A9VDP7 A0A0P5VNF0 A0A1A9ZSV6 A0A1A9Y0J0 A0A1B0B2Y9 A0A1Q3FZ93 A0A1A9W5Q7 A0A1Q3FZ63 B4LWB1 A0A0M4ER32 A0A0Q9WRK6 A0A0N8E7Q9 A0A182H4T2 A0A0P6I6T4 Q295F4 B4GDY8 A0A0P6CHJ2 B4JY64 A0A182H854
A0A1B3IIF8 A0A1B6ENC1 A0A2J7QMK8 A0A1B6CPI8 A0A067R1K6 A0A0A9W5Z2 A0A0K8SZ50 A0A1Y1ME68 A0A1Y1MCY1 A0A195EU65 A0A026WIX4 A0A195B0J4 A0A154PRQ2 A0A158NQW5 A0A195CM68 A0A195E5P9 F4WP72 A0A087ZZ67 A0A2A3EGS0 A0A232EZ17 A0A151WM91 E0VHE2 A0A224X5J2 N6T8T5 E2BBG0 A0A0C9QB16 A0A2S2Q4I6 A0A2H8TTG8 A0A2S2NZ57 X1XBF9 A0A139WNP4 A0A0M9A8L9 A0A139WNQ0 A0A310SDU4 W4VRM9 A0A3Q0IPR7 A0A3Q0JHC2 A0A336LRD9 A0A034VYW9 A0A0L0BR94 A0A1D2MXX3 W8B851 C4IXZ0 B4I3N0 A0A0A1WYZ4 A0A1I8NGE1 Q9VN14 A0A1W4VLC8 B4NID8 A0A1J1HHK2 A0A0P5RFZ7 A0A0N8E3P1 A0A1I8QBD3 A0A0P5MYZ4 B4K8Y9 A0A0N8BPK1 A0A0P6G4L4 A0A0P6H0T8 A0A0P6G5D7 A0A0P5S5M2 A0A0P5P8J5 A0A0P5QYP0 A0A0P5MLA3 A0A0P5JZS8 A0A0P5R4J0 A0A0P6D687 A0A0P4ZQ78 A0A1S4FFB1 A0A3B0KEU5 A0A0N8BQQ1 Q174B0 B4PTR6 A0A3B0JRG3 A0A0P4YSM5 B3LY10 A0A1A9VDP7 A0A0P5VNF0 A0A1A9ZSV6 A0A1A9Y0J0 A0A1B0B2Y9 A0A1Q3FZ93 A0A1A9W5Q7 A0A1Q3FZ63 B4LWB1 A0A0M4ER32 A0A0Q9WRK6 A0A0N8E7Q9 A0A182H4T2 A0A0P6I6T4 Q295F4 B4GDY8 A0A0P6CHJ2 B4JY64 A0A182H854
EC Number
6.2.1.4
Pubmed
26227816
18712529
26354079
27535920
24845553
25401762
+ More
26823975 28004739 24508170 30249741 21347285 21719571 28648823 20566863 23537049 20798317 18362917 19820115 25348373 26108605 27289101 24495485 17994087 25830018 25315136 15459097 10731132 12537572 12537569 17893096 19349973 18057021 17510324 17550304 26483478 15632085
26823975 28004739 24508170 30249741 21347285 21719571 28648823 20566863 23537049 20798317 18362917 19820115 25348373 26108605 27289101 24495485 17994087 25830018 25315136 15459097 10731132 12537572 12537569 17893096 19349973 18057021 17510324 17550304 26483478 15632085
EMBL
ODYU01004126
SOQ43672.1
JTDY01000914
KOB75456.1
NWSH01005469
PCG64178.1
+ More
AB264690 BAG30765.1 KQ459220 KPJ02908.1 RSAL01000222 RVE44025.1 KU840799 AOF39986.1 GECZ01030373 JAS39396.1 NEVH01013208 PNF29807.1 GEDC01022085 JAS15213.1 KK852812 KDR15864.1 GBHO01043325 GBHO01043324 GBHO01043323 GBHO01043322 GBHO01043321 GBHO01043320 GBHO01043319 GBHO01043318 GBHO01043317 GBHO01026697 GBHO01026696 GDHC01011104 GDHC01009280 JAG00279.1 JAG00280.1 JAG00281.1 JAG00282.1 JAG00283.1 JAG00284.1 JAG00285.1 JAG00286.1 JAG00287.1 JAG16907.1 JAG16908.1 JAQ07525.1 JAQ09349.1 GBRD01007631 JAG58190.1 GEZM01037123 JAV82156.1 GEZM01037124 JAV82155.1 KQ981965 KYN31788.1 KK107182 QOIP01000003 EZA55992.1 RLU25075.1 KQ976691 KYM77981.1 KQ435031 KZC14024.1 ADTU01023676 KQ977574 KYN01826.1 KQ979608 KYN20411.1 GL888243 EGI64003.1 KZ288262 PBC30356.1 NNAY01001615 OXU23418.1 KQ982944 KYQ48953.1 DS235169 EEB12798.1 GFTR01008680 JAW07746.1 APGK01047078 KB741077 KB632384 ENN74113.1 ERL94243.1 GL447043 EFN86967.1 GBYB01000424 GBYB01002675 JAG70191.1 JAG72442.1 GGMS01003433 MBY72636.1 GFXV01005738 MBW17543.1 GGMR01009836 MBY22455.1 ABLF02031408 KQ971307 KYB29620.1 KQ435711 KOX79460.1 KYB29619.1 KQ767242 OAD53473.1 GANO01001293 JAB58578.1 UFQS01000048 UFQT01000048 SSW98581.1 SSX18967.1 GAKP01011867 GAKP01011863 JAC47085.1 JRES01001482 KNC22536.1 LJIJ01000398 ODM97899.1 GAMC01011748 JAB94807.1 BT083421 ACQ91625.1 CH480821 EDW54823.1 GBXI01010562 JAD03730.1 AY229991 AE014297 BT021333 AY095040 CH964272 EDW83720.1 KRF99863.1 CVRI01000004 CRK87512.1 GDIQ01211082 GDIQ01169650 GDIQ01151413 GDIQ01120620 GDIQ01064939 GDIQ01064938 GDIQ01061732 GDIQ01030315 JAL31106.1 GDIQ01064940 GDIQ01047604 JAN29797.1 GDIQ01157452 JAK94273.1 CH933806 EDW16586.1 GDIQ01157451 JAK94274.1 GDIQ01038407 JAN56330.1 GDIQ01038408 JAN56329.1 GDIQ01211084 GDIQ01151362 GDIQ01049110 JAN45627.1 GDIQ01092632 JAL59094.1 GDIQ01138361 JAL13365.1 GDIQ01107968 JAL43758.1 GDIQ01154253 JAK97472.1 GDIQ01192945 JAK58780.1 GDIQ01105629 JAL46097.1 GDIQ01090178 JAN04559.1 GDIP01213910 JAJ09492.1 OUUW01000007 SPP83581.1 GDIQ01154251 JAK97474.1 CH477414 EAT41415.1 CM000160 EDW95649.1 SPP83583.1 GDIP01225061 JAI98340.1 CH902617 EDV42866.1 KPU79964.1 GDIP01097684 JAM06031.1 JXJN01007739 GFDL01002192 JAV32853.1 GFDL01002190 JAV32855.1 CH940650 EDW67645.2 CP012526 ALC46897.1 KRF83380.1 GDIQ01053596 JAN41141.1 JXUM01025782 KQ560733 KXJ81095.1 GDIQ01015848 JAN78889.1 CM000070 EAL28758.1 CH479182 EDW34649.1 GDIP01015021 JAM88694.1 CH916377 EDV90626.1 JXUM01028746 JXUM01028747 KQ560831 KXJ80691.1
AB264690 BAG30765.1 KQ459220 KPJ02908.1 RSAL01000222 RVE44025.1 KU840799 AOF39986.1 GECZ01030373 JAS39396.1 NEVH01013208 PNF29807.1 GEDC01022085 JAS15213.1 KK852812 KDR15864.1 GBHO01043325 GBHO01043324 GBHO01043323 GBHO01043322 GBHO01043321 GBHO01043320 GBHO01043319 GBHO01043318 GBHO01043317 GBHO01026697 GBHO01026696 GDHC01011104 GDHC01009280 JAG00279.1 JAG00280.1 JAG00281.1 JAG00282.1 JAG00283.1 JAG00284.1 JAG00285.1 JAG00286.1 JAG00287.1 JAG16907.1 JAG16908.1 JAQ07525.1 JAQ09349.1 GBRD01007631 JAG58190.1 GEZM01037123 JAV82156.1 GEZM01037124 JAV82155.1 KQ981965 KYN31788.1 KK107182 QOIP01000003 EZA55992.1 RLU25075.1 KQ976691 KYM77981.1 KQ435031 KZC14024.1 ADTU01023676 KQ977574 KYN01826.1 KQ979608 KYN20411.1 GL888243 EGI64003.1 KZ288262 PBC30356.1 NNAY01001615 OXU23418.1 KQ982944 KYQ48953.1 DS235169 EEB12798.1 GFTR01008680 JAW07746.1 APGK01047078 KB741077 KB632384 ENN74113.1 ERL94243.1 GL447043 EFN86967.1 GBYB01000424 GBYB01002675 JAG70191.1 JAG72442.1 GGMS01003433 MBY72636.1 GFXV01005738 MBW17543.1 GGMR01009836 MBY22455.1 ABLF02031408 KQ971307 KYB29620.1 KQ435711 KOX79460.1 KYB29619.1 KQ767242 OAD53473.1 GANO01001293 JAB58578.1 UFQS01000048 UFQT01000048 SSW98581.1 SSX18967.1 GAKP01011867 GAKP01011863 JAC47085.1 JRES01001482 KNC22536.1 LJIJ01000398 ODM97899.1 GAMC01011748 JAB94807.1 BT083421 ACQ91625.1 CH480821 EDW54823.1 GBXI01010562 JAD03730.1 AY229991 AE014297 BT021333 AY095040 CH964272 EDW83720.1 KRF99863.1 CVRI01000004 CRK87512.1 GDIQ01211082 GDIQ01169650 GDIQ01151413 GDIQ01120620 GDIQ01064939 GDIQ01064938 GDIQ01061732 GDIQ01030315 JAL31106.1 GDIQ01064940 GDIQ01047604 JAN29797.1 GDIQ01157452 JAK94273.1 CH933806 EDW16586.1 GDIQ01157451 JAK94274.1 GDIQ01038407 JAN56330.1 GDIQ01038408 JAN56329.1 GDIQ01211084 GDIQ01151362 GDIQ01049110 JAN45627.1 GDIQ01092632 JAL59094.1 GDIQ01138361 JAL13365.1 GDIQ01107968 JAL43758.1 GDIQ01154253 JAK97472.1 GDIQ01192945 JAK58780.1 GDIQ01105629 JAL46097.1 GDIQ01090178 JAN04559.1 GDIP01213910 JAJ09492.1 OUUW01000007 SPP83581.1 GDIQ01154251 JAK97474.1 CH477414 EAT41415.1 CM000160 EDW95649.1 SPP83583.1 GDIP01225061 JAI98340.1 CH902617 EDV42866.1 KPU79964.1 GDIP01097684 JAM06031.1 JXJN01007739 GFDL01002192 JAV32853.1 GFDL01002190 JAV32855.1 CH940650 EDW67645.2 CP012526 ALC46897.1 KRF83380.1 GDIQ01053596 JAN41141.1 JXUM01025782 KQ560733 KXJ81095.1 GDIQ01015848 JAN78889.1 CM000070 EAL28758.1 CH479182 EDW34649.1 GDIP01015021 JAM88694.1 CH916377 EDV90626.1 JXUM01028746 JXUM01028747 KQ560831 KXJ80691.1
Proteomes
UP000037510
UP000218220
UP000053268
UP000283053
UP000235965
UP000027135
+ More
UP000078541 UP000053097 UP000279307 UP000078540 UP000076502 UP000005205 UP000078542 UP000078492 UP000007755 UP000005203 UP000242457 UP000215335 UP000075809 UP000009046 UP000019118 UP000030742 UP000008237 UP000007819 UP000007266 UP000053105 UP000079169 UP000037069 UP000094527 UP000001292 UP000095301 UP000000803 UP000192221 UP000007798 UP000183832 UP000095300 UP000009192 UP000268350 UP000008820 UP000002282 UP000007801 UP000078200 UP000092445 UP000092443 UP000092460 UP000091820 UP000008792 UP000092553 UP000069940 UP000249989 UP000001819 UP000008744 UP000001070
UP000078541 UP000053097 UP000279307 UP000078540 UP000076502 UP000005205 UP000078542 UP000078492 UP000007755 UP000005203 UP000242457 UP000215335 UP000075809 UP000009046 UP000019118 UP000030742 UP000008237 UP000007819 UP000007266 UP000053105 UP000079169 UP000037069 UP000094527 UP000001292 UP000095301 UP000000803 UP000192221 UP000007798 UP000183832 UP000095300 UP000009192 UP000268350 UP000008820 UP000002282 UP000007801 UP000078200 UP000092445 UP000092443 UP000092460 UP000091820 UP000008792 UP000092553 UP000069940 UP000249989 UP000001819 UP000008744 UP000001070
Pfam
Interpro
IPR036770
Ankyrin_rpt-contain_sf
+ More
IPR013098 Ig_I-set
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR020683 Ankyrin_rpt-contain_dom
IPR016187 CTDL_fold
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR036116 FN3_sf
IPR001304 C-type_lectin-like
IPR002110 Ankyrin_rpt
IPR003961 FN3_dom
IPR016186 C-type_lectin-like/link_sf
IPR007110 Ig-like_dom
IPR008646 Herpes_UL45-like
IPR016162 Ald_DH_N
IPR010061 MeMal-semiAld_DH
IPR015590 Aldehyde_DH_dom
IPR016163 Ald_DH_C
IPR016160 Ald_DH_CS_CYS
IPR016161 Ald_DH/histidinol_DH
IPR034722 Succ_CoA_betaG_euk
IPR013815 ATP_grasp_subdomain_1
IPR017866 Succ-CoA_synthase_bsu_CS
IPR005809 Succ_CoA_synthase_bsu
IPR013650 ATP-grasp_succ-CoA_synth-type
IPR005811 CoA_ligase
IPR016102 Succinyl-CoA_synth-like
IPR003529 Hematopoietin_rcpt_Gp130_CS
IPR017986 WD40_repeat_dom
IPR024977 Apc4_WD40_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR015048 DUF1899
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR027331 CORO7
IPR015505 Coronin
IPR013151 Immunoglobulin
IPR013098 Ig_I-set
IPR013783 Ig-like_fold
IPR003599 Ig_sub
IPR020683 Ankyrin_rpt-contain_dom
IPR016187 CTDL_fold
IPR036179 Ig-like_dom_sf
IPR003598 Ig_sub2
IPR036116 FN3_sf
IPR001304 C-type_lectin-like
IPR002110 Ankyrin_rpt
IPR003961 FN3_dom
IPR016186 C-type_lectin-like/link_sf
IPR007110 Ig-like_dom
IPR008646 Herpes_UL45-like
IPR016162 Ald_DH_N
IPR010061 MeMal-semiAld_DH
IPR015590 Aldehyde_DH_dom
IPR016163 Ald_DH_C
IPR016160 Ald_DH_CS_CYS
IPR016161 Ald_DH/histidinol_DH
IPR034722 Succ_CoA_betaG_euk
IPR013815 ATP_grasp_subdomain_1
IPR017866 Succ-CoA_synthase_bsu_CS
IPR005809 Succ_CoA_synthase_bsu
IPR013650 ATP-grasp_succ-CoA_synth-type
IPR005811 CoA_ligase
IPR016102 Succinyl-CoA_synth-like
IPR003529 Hematopoietin_rcpt_Gp130_CS
IPR017986 WD40_repeat_dom
IPR024977 Apc4_WD40_dom
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR015048 DUF1899
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR027331 CORO7
IPR015505 Coronin
IPR013151 Immunoglobulin
SUPFAM
Gene 3D
ProteinModelPortal
A0A2H1VT68
A0A0L7LJS0
A0A2A4IXP8
B2DBK6
A0A194QBI6
A0A3S2LTD0
+ More
A0A1B3IIF8 A0A1B6ENC1 A0A2J7QMK8 A0A1B6CPI8 A0A067R1K6 A0A0A9W5Z2 A0A0K8SZ50 A0A1Y1ME68 A0A1Y1MCY1 A0A195EU65 A0A026WIX4 A0A195B0J4 A0A154PRQ2 A0A158NQW5 A0A195CM68 A0A195E5P9 F4WP72 A0A087ZZ67 A0A2A3EGS0 A0A232EZ17 A0A151WM91 E0VHE2 A0A224X5J2 N6T8T5 E2BBG0 A0A0C9QB16 A0A2S2Q4I6 A0A2H8TTG8 A0A2S2NZ57 X1XBF9 A0A139WNP4 A0A0M9A8L9 A0A139WNQ0 A0A310SDU4 W4VRM9 A0A3Q0IPR7 A0A3Q0JHC2 A0A336LRD9 A0A034VYW9 A0A0L0BR94 A0A1D2MXX3 W8B851 C4IXZ0 B4I3N0 A0A0A1WYZ4 A0A1I8NGE1 Q9VN14 A0A1W4VLC8 B4NID8 A0A1J1HHK2 A0A0P5RFZ7 A0A0N8E3P1 A0A1I8QBD3 A0A0P5MYZ4 B4K8Y9 A0A0N8BPK1 A0A0P6G4L4 A0A0P6H0T8 A0A0P6G5D7 A0A0P5S5M2 A0A0P5P8J5 A0A0P5QYP0 A0A0P5MLA3 A0A0P5JZS8 A0A0P5R4J0 A0A0P6D687 A0A0P4ZQ78 A0A1S4FFB1 A0A3B0KEU5 A0A0N8BQQ1 Q174B0 B4PTR6 A0A3B0JRG3 A0A0P4YSM5 B3LY10 A0A1A9VDP7 A0A0P5VNF0 A0A1A9ZSV6 A0A1A9Y0J0 A0A1B0B2Y9 A0A1Q3FZ93 A0A1A9W5Q7 A0A1Q3FZ63 B4LWB1 A0A0M4ER32 A0A0Q9WRK6 A0A0N8E7Q9 A0A182H4T2 A0A0P6I6T4 Q295F4 B4GDY8 A0A0P6CHJ2 B4JY64 A0A182H854
A0A1B3IIF8 A0A1B6ENC1 A0A2J7QMK8 A0A1B6CPI8 A0A067R1K6 A0A0A9W5Z2 A0A0K8SZ50 A0A1Y1ME68 A0A1Y1MCY1 A0A195EU65 A0A026WIX4 A0A195B0J4 A0A154PRQ2 A0A158NQW5 A0A195CM68 A0A195E5P9 F4WP72 A0A087ZZ67 A0A2A3EGS0 A0A232EZ17 A0A151WM91 E0VHE2 A0A224X5J2 N6T8T5 E2BBG0 A0A0C9QB16 A0A2S2Q4I6 A0A2H8TTG8 A0A2S2NZ57 X1XBF9 A0A139WNP4 A0A0M9A8L9 A0A139WNQ0 A0A310SDU4 W4VRM9 A0A3Q0IPR7 A0A3Q0JHC2 A0A336LRD9 A0A034VYW9 A0A0L0BR94 A0A1D2MXX3 W8B851 C4IXZ0 B4I3N0 A0A0A1WYZ4 A0A1I8NGE1 Q9VN14 A0A1W4VLC8 B4NID8 A0A1J1HHK2 A0A0P5RFZ7 A0A0N8E3P1 A0A1I8QBD3 A0A0P5MYZ4 B4K8Y9 A0A0N8BPK1 A0A0P6G4L4 A0A0P6H0T8 A0A0P6G5D7 A0A0P5S5M2 A0A0P5P8J5 A0A0P5QYP0 A0A0P5MLA3 A0A0P5JZS8 A0A0P5R4J0 A0A0P6D687 A0A0P4ZQ78 A0A1S4FFB1 A0A3B0KEU5 A0A0N8BQQ1 Q174B0 B4PTR6 A0A3B0JRG3 A0A0P4YSM5 B3LY10 A0A1A9VDP7 A0A0P5VNF0 A0A1A9ZSV6 A0A1A9Y0J0 A0A1B0B2Y9 A0A1Q3FZ93 A0A1A9W5Q7 A0A1Q3FZ63 B4LWB1 A0A0M4ER32 A0A0Q9WRK6 A0A0N8E7Q9 A0A182H4T2 A0A0P6I6T4 Q295F4 B4GDY8 A0A0P6CHJ2 B4JY64 A0A182H854
PDB
5I99
E-value=3.47949e-59,
Score=583
Ontologies
GO
GO:0016021
GO:0004491
GO:0006099
GO:0005525
GO:0006104
GO:0005524
GO:0000287
GO:0005739
GO:0004776
GO:0004775
GO:0016020
GO:0004896
GO:0005794
GO:0003824
GO:0019991
GO:0061343
GO:0021682
GO:0008366
GO:0060857
GO:0005918
GO:0031225
GO:0045197
GO:0005886
GO:0005515
GO:0004222
GO:0006508
GO:0003677
GO:0003887
GO:0001664
GO:0016491
PANTHER
Topology
Subcellular location
Mitochondrion
Cell membrane
Cell junction
Septate junction
Cell membrane
Cell junction
Septate junction
SignalP
Position: 1 - 16,
Likelihood: 0.994938
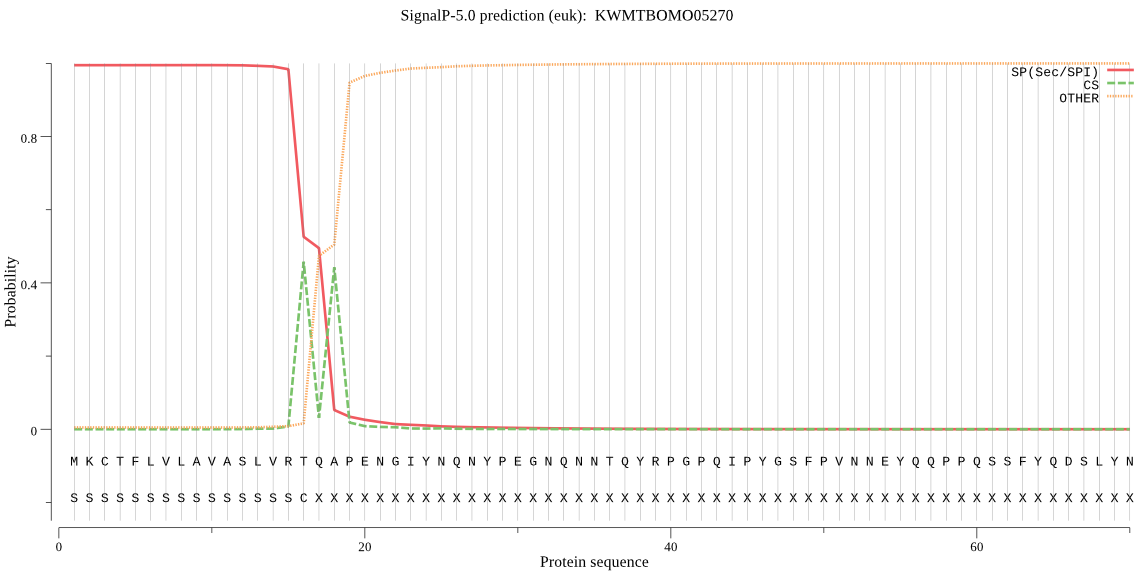
Length:
1142
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01393
Exp number, first 60 AAs:
0.00134
Total prob of N-in:
0.00008
outside
1 - 1142
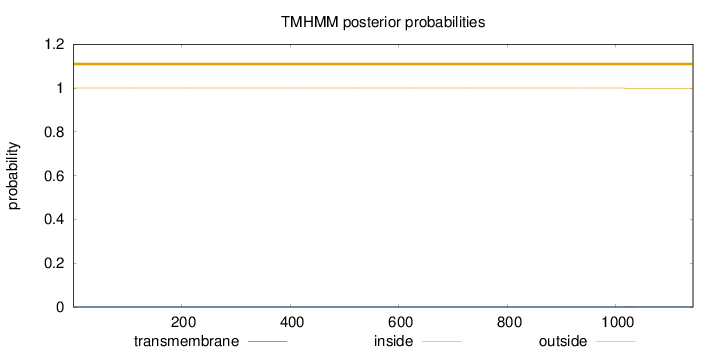
Population Genetic Test Statistics
Pi
202.330612
Theta
183.265772
Tajima's D
0.270979
CLR
0.087149
CSRT
0.451127443627819
Interpretation
Uncertain
