Gene
KWMTBOMO05267
Pre Gene Modal
BGIBMGA012813
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.674
Sequence
CDS
ATGTTGATTAAATTAGAAGCCAAACCATTTAACATCAACATTGTCCAAGTTTATGCGCCTACAGGGGATAAACCAATGGGCCAGATAGAGGACTTATATAGGGCTATTGATAGCTTATTAAAATCTACTAAGAATCACGAGTTAGTTATCGTTATGGGTGACATGAATGCGAAGGTGGGTTCGACCATTACTCCTGGAGTGACTGGTGCTTATGGCCTTGGAAACAGAAACGATAGAGATCATAACCCAGTGGTGGTAGACGTAGCATGCAAGCTGAAGAAGTTGCAGCCAAAGAGAATGGATGGAAGGCCAAACGTACGGAAGCTTGCTGAGCCCGACACACGAGCGAAGACCCACACCGCTATTAATCAATGGGCCAACGATCGTCGCACTGAAACCACTGCCTCTGACCTCGACACGTGGACTAGTCTAAAGCGAAGTGTAAAAGATATATGTCACCAATACCTCCAACCATCACAACTGGTCAAAAAACAAGATTGGATGACAGATTCAATTTTGCAGCTTATGGAAGACAGGAGGCAATTTAAGGGTACAAATCCAGTAATGTATAAGCAAATCGATGTCACCATTCGAAAAGAGATTGCGAAAGCCAAGAATGATTACTACCGCAATAAGTGCGCGCACTTGGAAGATCTCCTTCGTAAACATGACAGCTTCAATGCGCACAAGCACATCAAGGAAATGGTTGGTCATAGACGGAAATCAATGAATACTCTGACCGATGCAGGTGGGAACCTCATTCTTGACACCGATGGCAAAAAGAGAGTGTGGTGTGACTATGTTGCAGAGATGTTTGCGGACTCATCGAGGAACATTTCACCGATCACTGATCGGACTGGTCCAGCTATGTTGCGATCTGAGGTTTTGCGGGCCTTGAAAGCTGCTAAAACAGGTAAATCACCGGGTCCTGATAATATACCGATCGAAATACTAAAGTTGCTGGAAGAAGATCAACTGGACGTTCTCACACAGTTCTTCAACAACATCTACGAAACAGGCAATCTTCCTAGTGACTGGCTAAAATCCACATTCATTACGATACCCAAGAAGCAAAATGCCAAAAAATGTGGAGAGTATCGTATGATTAGTCTGATGAGCCATGTCCTAAAAGTATTCTTGAACATCATACAGAATAGAATACGGCCAAAATGTGATGAACAACTGGGCGATAGCCAATTTGGATTTCGATCTGGTGTTGGCACCAGAGAAGCTCTTTTCGCTCTCAACGTTCTCGTACAGAAATGCAGAGACATGCAAACTGATGTCTTTTTGTGCTTCATAGACTACGAAAAGGCATTTGACCGAGTTAAACATCATCAACTTTTTAGCCTTTTATGTGACATTGGTCTAGATGGTAAGGACGTCAGAATCATCCGCAACCTGTACGAGAAGCAGGTGGCTACGATAAGAGTCGAAAATGAGGAAACCGACCAAGTTGAGATCTGTCGTGGGGTCCGGCAGGGAAGCACGAAGGATCGGAATTTTCCTGTTAAGAAACAAAAGCGTACTCGCTTCGGTGTGAAAAGCAGGGAGATTCTGGCCCAAATTGTTTCGTAG
Protein
MLIKLEAKPFNINIVQVYAPTGDKPMGQIEDLYRAIDSLLKSTKNHELVIVMGDMNAKVGSTITPGVTGAYGLGNRNDRDHNPVVVDVACKLKKLQPKRMDGRPNVRKLAEPDTRAKTHTAINQWANDRRTETTASDLDTWTSLKRSVKDICHQYLQPSQLVKKQDWMTDSILQLMEDRRQFKGTNPVMYKQIDVTIRKEIAKAKNDYYRNKCAHLEDLLRKHDSFNAHKHIKEMVGHRRKSMNTLTDAGGNLILDTDGKKRVWCDYVAEMFADSSRNISPITDRTGPAMLRSEVLRALKAAKTGKSPGPDNIPIEILKLLEEDQLDVLTQFFNNIYETGNLPSDWLKSTFITIPKKQNAKKCGEYRMISLMSHVLKVFLNIIQNRIRPKCDEQLGDSQFGFRSGVGTREALFALNVLVQKCRDMQTDVFLCFIDYEKAFDRVKHHQLFSLLCDIGLDGKDVRIIRNLYEKQVATIRVENEETDQVEICRGVRQGSTKDRNFPVKKQKRTRFGVKSREILAQIVS
Summary
Uniprot
D5LB38
A0A2A4JBK9
A0A3S1C192
A0A2V9X8L4
A0A0B7BU85
A0A0B7BRW7
+ More
A0A0B7BTM4 A0A0B7BU90 A0A0B7BR36 A0A0B7BRV7 A0A1W7RAL1 A0A076N1B6 A0A085NA39 A0A023GD30 A0A0B1PP52 A0A2J7QCR1 A0A2A4JXP9 A0A2J7PCW0 A0A2J7PWT0 A0A2J7RE76 A0A2J7QPT5 A0A076N3Z4 A0A2J7PMY4 A0A2J7QLQ1 A0A2S2PMS6 A0A2J7PHX9 A0A2J7Q6F7 A0A2J7R7D2 A0A2J7PTB8 A0A2J7Q7C6 A0A2J7PFC5 A0A2J7PBF1 A0A2J7PSI0 A0A2J7RL19 A0A2J7PC89 A0A2J7Q6P7 A0A2J7QCG2 A0A2J7PSH5 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7QNQ3 A0A2J7RJY3 A0A2J7R8P5 A0A2J7QI56 A0A2J7PNH4 A0A2J7RRE4 A0A2J7QMC5 A0A2J7PZN7 A0A2J7PT27 A0A2J7RBL1 A0A2J7PF68 A0A2J7RPX5 A0A2J7RBL5 A0A2J7Q649 A0A2J7QSZ5 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RNM5 A0A2J7QJJ9 A0A146LUY8 A0A2J7QEJ0 A0A2J7PTY0 A0A2J7R901 A0A2J7RIL8 A0A2J7QR01 A0A2J7PW81 H3ABE5 A0A2J7PQA3 A0A2J7REE7 A0A2J7PVW0 A0A2J7RLI7 A0A2J7PX03 A0A2J7RP47 A0A2J7PMJ3 A0A2J7RGX2 A0A2J7QGX8 A0A2J7R639 A0A2J7Q0C6 A0A2J7QGT4 A0A2J7QH51
A0A0B7BTM4 A0A0B7BU90 A0A0B7BR36 A0A0B7BRV7 A0A1W7RAL1 A0A076N1B6 A0A085NA39 A0A023GD30 A0A0B1PP52 A0A2J7QCR1 A0A2A4JXP9 A0A2J7PCW0 A0A2J7PWT0 A0A2J7RE76 A0A2J7QPT5 A0A076N3Z4 A0A2J7PMY4 A0A2J7QLQ1 A0A2S2PMS6 A0A2J7PHX9 A0A2J7Q6F7 A0A2J7R7D2 A0A2J7PTB8 A0A2J7Q7C6 A0A2J7PFC5 A0A2J7PBF1 A0A2J7PSI0 A0A2J7RL19 A0A2J7PC89 A0A2J7Q6P7 A0A2J7QCG2 A0A2J7PSH5 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7QNQ3 A0A2J7RJY3 A0A2J7R8P5 A0A2J7QI56 A0A2J7PNH4 A0A2J7RRE4 A0A2J7QMC5 A0A2J7PZN7 A0A2J7PT27 A0A2J7RBL1 A0A2J7PF68 A0A2J7RPX5 A0A2J7RBL5 A0A2J7Q649 A0A2J7QSZ5 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RNM5 A0A2J7QJJ9 A0A146LUY8 A0A2J7QEJ0 A0A2J7PTY0 A0A2J7R901 A0A2J7RIL8 A0A2J7QR01 A0A2J7PW81 H3ABE5 A0A2J7PQA3 A0A2J7REE7 A0A2J7PVW0 A0A2J7RLI7 A0A2J7PX03 A0A2J7RP47 A0A2J7PMJ3 A0A2J7RGX2 A0A2J7QGX8 A0A2J7R639 A0A2J7Q0C6 A0A2J7QGT4 A0A2J7QH51
EMBL
GU815089
ADF18552.1
NWSH01001998
PCG69477.1
RQTK01000404
RUS80216.1
+ More
QIAP01000240 PYX61602.1 HACG01048870 CEK95735.1 HACG01048883 CEK95748.1 HACG01048876 CEK95741.1 HACG01048875 CEK95740.1 HACG01048879 CEK95744.1 HACG01048873 CEK95738.1 GFAH01000222 JAV48167.1 KF881087 AIJ27486.1 KL367526 KFD66335.1 GBBM01003639 JAC31779.1 KN538403 KHJ42451.1 NEVH01016127 PNF26367.1 NWSH01000469 PCG76200.1 NEVH01026403 PNF14170.1 NEVH01020868 PNF20797.1 NEVH01005277 PNF39130.1 NEVH01012087 PNF30599.1 KF881086 AIJ27485.1 NEVH01023960 PNF17698.1 NEVH01013243 PNF29503.1 GGMR01018096 MBY30715.1 NEVH01025130 PNF15934.1 NEVH01017478 PNF24170.1 NEVH01006734 PNF36732.1 NEVH01021610 PNF19569.1 NEVH01017443 PNF24485.1 NEVH01025668 PNF15041.1 NEVH01027104 PNF13657.1 NEVH01021928 PNF19298.1 NEVH01002700 PNF41535.1 NEVH01027067 PNF13944.1 NEVH01017460 PNF24248.1 NEVH01016289 PNF26277.1 NEVH01021931 PNF19256.1 NEVH01017452 PNF24293.1 NEVH01019066 PNF23173.1 NEVH01012564 PNF30212.1 NEVH01002982 PNF41147.1 NEVH01006721 PNF37211.1 NEVH01013959 PNF28275.1 NEVH01023953 PNF17877.1 NEVH01000609 PNF43402.1 NEVH01013215 PNF29703.1 NEVH01020333 PNF21790.1 NEVH01021921 PNF19488.1 NEVH01005896 PNF38208.1 NEVH01026085 PNF14968.1 NEVH01020851 NEVH01013954 NEVH01006574 NEVH01001358 NEVH01001354 PNF21237.1 PNF28332.1 PNF37782.1 PNF42792.1 PNF42884.1 PNF38212.1 NEVH01017540 PNF24056.1 NEVH01011205 PNF31711.1 NEVH01011186 PNF32183.1 NEVH01016335 PNF25462.1 PNF37115.1 NEVH01002150 PNF42436.1 NEVH01013553 PNF28765.1 GDHC01007098 GDHC01001590 JAQ11531.1 JAQ17039.1 NEVH01015309 PNF27009.1 NEVH01021219 PNF19787.1 PNF37310.1 NEVH01003500 PNF40666.1 NEVH01012082 PNF31006.1 NEVH01020937 PNF20581.1 AFYH01236276 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 PNF39212.1 NEVH01020940 PNF20478.1 NEVH01002684 PNF41704.1 NEVH01020865 PNF20863.1 NEVH01002141 PNF42599.1 NEVH01023985 PNF17546.1 NEVH01003750 PNF40091.1 NEVH01014359 PNF27813.1 NEVH01006981 PNF36299.1 NEVH01019971 PNF22037.1 PNF27789.1 NEVH01014358 PNF27914.1
QIAP01000240 PYX61602.1 HACG01048870 CEK95735.1 HACG01048883 CEK95748.1 HACG01048876 CEK95741.1 HACG01048875 CEK95740.1 HACG01048879 CEK95744.1 HACG01048873 CEK95738.1 GFAH01000222 JAV48167.1 KF881087 AIJ27486.1 KL367526 KFD66335.1 GBBM01003639 JAC31779.1 KN538403 KHJ42451.1 NEVH01016127 PNF26367.1 NWSH01000469 PCG76200.1 NEVH01026403 PNF14170.1 NEVH01020868 PNF20797.1 NEVH01005277 PNF39130.1 NEVH01012087 PNF30599.1 KF881086 AIJ27485.1 NEVH01023960 PNF17698.1 NEVH01013243 PNF29503.1 GGMR01018096 MBY30715.1 NEVH01025130 PNF15934.1 NEVH01017478 PNF24170.1 NEVH01006734 PNF36732.1 NEVH01021610 PNF19569.1 NEVH01017443 PNF24485.1 NEVH01025668 PNF15041.1 NEVH01027104 PNF13657.1 NEVH01021928 PNF19298.1 NEVH01002700 PNF41535.1 NEVH01027067 PNF13944.1 NEVH01017460 PNF24248.1 NEVH01016289 PNF26277.1 NEVH01021931 PNF19256.1 NEVH01017452 PNF24293.1 NEVH01019066 PNF23173.1 NEVH01012564 PNF30212.1 NEVH01002982 PNF41147.1 NEVH01006721 PNF37211.1 NEVH01013959 PNF28275.1 NEVH01023953 PNF17877.1 NEVH01000609 PNF43402.1 NEVH01013215 PNF29703.1 NEVH01020333 PNF21790.1 NEVH01021921 PNF19488.1 NEVH01005896 PNF38208.1 NEVH01026085 PNF14968.1 NEVH01020851 NEVH01013954 NEVH01006574 NEVH01001358 NEVH01001354 PNF21237.1 PNF28332.1 PNF37782.1 PNF42792.1 PNF42884.1 PNF38212.1 NEVH01017540 PNF24056.1 NEVH01011205 PNF31711.1 NEVH01011186 PNF32183.1 NEVH01016335 PNF25462.1 PNF37115.1 NEVH01002150 PNF42436.1 NEVH01013553 PNF28765.1 GDHC01007098 GDHC01001590 JAQ11531.1 JAQ17039.1 NEVH01015309 PNF27009.1 NEVH01021219 PNF19787.1 PNF37310.1 NEVH01003500 PNF40666.1 NEVH01012082 PNF31006.1 NEVH01020937 PNF20581.1 AFYH01236276 NEVH01022640 NEVH01008218 NEVH01005885 PNF18513.1 PNF34430.1 PNF38481.1 PNF39212.1 NEVH01020940 PNF20478.1 NEVH01002684 PNF41704.1 NEVH01020865 PNF20863.1 NEVH01002141 PNF42599.1 NEVH01023985 PNF17546.1 NEVH01003750 PNF40091.1 NEVH01014359 PNF27813.1 NEVH01006981 PNF36299.1 NEVH01019971 PNF22037.1 PNF27789.1 NEVH01014358 PNF27914.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
D5LB38
A0A2A4JBK9
A0A3S1C192
A0A2V9X8L4
A0A0B7BU85
A0A0B7BRW7
+ More
A0A0B7BTM4 A0A0B7BU90 A0A0B7BR36 A0A0B7BRV7 A0A1W7RAL1 A0A076N1B6 A0A085NA39 A0A023GD30 A0A0B1PP52 A0A2J7QCR1 A0A2A4JXP9 A0A2J7PCW0 A0A2J7PWT0 A0A2J7RE76 A0A2J7QPT5 A0A076N3Z4 A0A2J7PMY4 A0A2J7QLQ1 A0A2S2PMS6 A0A2J7PHX9 A0A2J7Q6F7 A0A2J7R7D2 A0A2J7PTB8 A0A2J7Q7C6 A0A2J7PFC5 A0A2J7PBF1 A0A2J7PSI0 A0A2J7RL19 A0A2J7PC89 A0A2J7Q6P7 A0A2J7QCG2 A0A2J7PSH5 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7QNQ3 A0A2J7RJY3 A0A2J7R8P5 A0A2J7QI56 A0A2J7PNH4 A0A2J7RRE4 A0A2J7QMC5 A0A2J7PZN7 A0A2J7PT27 A0A2J7RBL1 A0A2J7PF68 A0A2J7RPX5 A0A2J7RBL5 A0A2J7Q649 A0A2J7QSZ5 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RNM5 A0A2J7QJJ9 A0A146LUY8 A0A2J7QEJ0 A0A2J7PTY0 A0A2J7R901 A0A2J7RIL8 A0A2J7QR01 A0A2J7PW81 H3ABE5 A0A2J7PQA3 A0A2J7REE7 A0A2J7PVW0 A0A2J7RLI7 A0A2J7PX03 A0A2J7RP47 A0A2J7PMJ3 A0A2J7RGX2 A0A2J7QGX8 A0A2J7R639 A0A2J7Q0C6 A0A2J7QGT4 A0A2J7QH51
A0A0B7BTM4 A0A0B7BU90 A0A0B7BR36 A0A0B7BRV7 A0A1W7RAL1 A0A076N1B6 A0A085NA39 A0A023GD30 A0A0B1PP52 A0A2J7QCR1 A0A2A4JXP9 A0A2J7PCW0 A0A2J7PWT0 A0A2J7RE76 A0A2J7QPT5 A0A076N3Z4 A0A2J7PMY4 A0A2J7QLQ1 A0A2S2PMS6 A0A2J7PHX9 A0A2J7Q6F7 A0A2J7R7D2 A0A2J7PTB8 A0A2J7Q7C6 A0A2J7PFC5 A0A2J7PBF1 A0A2J7PSI0 A0A2J7RL19 A0A2J7PC89 A0A2J7Q6P7 A0A2J7QCG2 A0A2J7PSH5 A0A2J7Q6T5 A0A2J7Q3K9 A0A2J7QNQ3 A0A2J7RJY3 A0A2J7R8P5 A0A2J7QI56 A0A2J7PNH4 A0A2J7RRE4 A0A2J7QMC5 A0A2J7PZN7 A0A2J7PT27 A0A2J7RBL1 A0A2J7PF68 A0A2J7RPX5 A0A2J7RBL5 A0A2J7Q649 A0A2J7QSZ5 A0A2J7QUD0 A0A2J7R8G1 A0A2J7RNM5 A0A2J7QJJ9 A0A146LUY8 A0A2J7QEJ0 A0A2J7PTY0 A0A2J7R901 A0A2J7RIL8 A0A2J7QR01 A0A2J7PW81 H3ABE5 A0A2J7PQA3 A0A2J7REE7 A0A2J7PVW0 A0A2J7RLI7 A0A2J7PX03 A0A2J7RP47 A0A2J7PMJ3 A0A2J7RGX2 A0A2J7QGX8 A0A2J7R639 A0A2J7Q0C6 A0A2J7QGT4 A0A2J7QH51
PDB
6AR3
E-value=0.00917949,
Score=93
Ontologies
KEGG
PANTHER
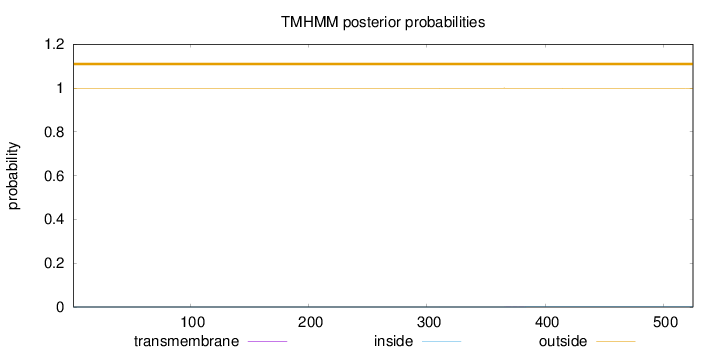
Topology
Length:
525
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00797
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.00049
outside
1 - 525
Population Genetic Test Statistics
Pi
329.724445
Theta
218.01055
Tajima's D
1.683661
CLR
0.095983
CSRT
0.825808709564522
Interpretation
Uncertain
