Gene
KWMTBOMO05263
Pre Gene Modal
BGIBMGA002495
Annotation
PREDICTED:_protein_LMBR1L_isoform_X2_[Bombyx_mori]
Full name
Protein Lilipod
Location in the cell
PlasmaMembrane Reliability : 4.955
Sequence
CDS
ATGGATGATGAAGAGGCAGATTTACGGGAACAGATTTTTCATAATAATGTTAGAGAACAAATTATTTTCTTGTTATTATTTATATTGCTGTATCTGATCTCGTTCACGTTGATTGAGAAGTTTCGGAGACGAGACAGCGAAGACTATTTTACAGCAGACGAAGATGAAGTCAAAGTGTACAGGATCAGCTTGTGGCTCTGCACATTTGCCTTGGCAGTGTCCCTGGGCTCGGCACTCCTGCTGCCCGTTTCGATTGTCAGCAATGAAGTGCTCATACTGTACCCAAACAGTTATTATGTAAAATGGCTCAATAGCTCTCTGATACAAGGTCTATGGAATCACGTATTTTTATTTTCAAATCTATCATTGTTTGTTTTCCTACCGTTTGCCTATTTATTTTCGGAGTCGTCCGGATTTCCTGGATGCCGCGGTCTGAAGGGTCGAGTCTACGAAACTTTTATTGTGCTAGCATTGCTCGGCATCGCTATGCTCGGCTTGGCCTATGTCATATCGGCTTGGATAGAAGGAGATCGTTCTGGTGTGGACGCATTATTGAATCTATGGACTTATCTACCGTTCCTCTATTCCTGCGTGTCTTTTGTCGGCGTTTTAATGTTACTAGTTTTCACGCCGCTTGGCTTCGTGCGTCTGTTTGGAGTTGTCGGCGGAGTTCTCGTGAAGCCGCAGTTCCTCAGAGATCTCAACGAGGAGTATTACATGTATTCGTTCGAAGAAGAGACAATCCGACGGCGAATCGACAATGCCGTCGCAACTGAATGA
Protein
MDDEEADLREQIFHNNVREQIIFLLLFILLYLISFTLIEKFRRRDSEDYFTADEDEVKVYRISLWLCTFALAVSLGSALLLPVSIVSNEVLILYPNSYYVKWLNSSLIQGLWNHVFLFSNLSLFVFLPFAYLFSESSGFPGCRGLKGRVYETFIVLALLGIAMLGLAYVISAWIEGDRSGVDALLNLWTYLPFLYSCVSFVGVLMLLVFTPLGFVRLFGVVGGVLVKPQFLRDLNEEYYMYSFEEETIRRRIDNAVATE
Summary
Description
Required during oogenesis to promote self-renewal of germline stem cells, probably by enhancing BMP signaling activity.
Similarity
Belongs to the LIMR family.
Keywords
Cell membrane
Complete proteome
Differentiation
Membrane
Oogenesis
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Protein Lilipod
Uniprot
A0A3S2NN29
A0A212F1E0
A0A1E1WKM5
A0A194RMG5
A0A194QBV4
S4PXJ0
+ More
A0A1W7R8U3 A0A2H1WJ69 A0A2J7RAF3 A0A087ZQT2 V5I8M1 V5GR09 A0A026WI55 A0A1Q3G2A0 A0A139WB21 Q16UA7 D6X3S1 A0A1Q3FX73 A0A0C9R641 A0A0P6K0Y0 A0A023EVE3 A0A1Q3G2B7 A0A1Y1MNM7 A0A1Q3FX89 A0A151WQ01 F4X5V0 A0A084W692 A0A154PLC9 A0A067RPE4 A0A2M4AQ35 A0A2M4AEX4 A0A034VJS4 A0A2M4AEQ0 A0A2A3EK57 A0A0A1X4X1 A0A2M4ADW3 A0A1L8E385 A0A0K8U1Q9 A0A0K8TMV1 W8CB69 A0A2M4CV67 A0A2M4CUX5 A0A1B0G2Z2 E9IEG0 A0A151JM01 A0A1A9X1P2 A0A336LSQ6 A0A336MNJ6 A0A151JVK9 A0A1B6CFN2 A0A0L7RCA1 A0A195CDY2 A0A1I8NTD7 A0A1I8NTD9 E2B7S6 A0A182QZ66 Q7QII1 A0A1B6MKD5 A0A1I8ME42 A0A1I8ME39 K7IXZ4 A0A1B6IDQ5 A0A336LN84 A0A146M6N0 A0A1B6K5F8 A0A1A9UQW6 B4NGP5 A0A0A9XLD6 A0A0T6B7K0 E2A3U5 A0A1B0BTE7 A0A069DU90 A0A182Y833 A0A182PIP7 A0A182RJE7 A0A182U2D9 A0A182I6K0 A0A182L3V4 A0A182VMB5 A0A182K2D1 A0A2M4BJ10 A0A1A9XK02 A0A1A9ZWR5 A0A023F450 R7U746 A0A1B0CCI3 A0A182FZP5 A0A1S3K926 B4HHK5 B4K5I6 V5H9U6 A0A182J7T1 B4JIK8 A0A1C9TA51 B4LWE0 A0A0B4KGZ8 B3LYA8 Q9VC35 B4QTS6
A0A1W7R8U3 A0A2H1WJ69 A0A2J7RAF3 A0A087ZQT2 V5I8M1 V5GR09 A0A026WI55 A0A1Q3G2A0 A0A139WB21 Q16UA7 D6X3S1 A0A1Q3FX73 A0A0C9R641 A0A0P6K0Y0 A0A023EVE3 A0A1Q3G2B7 A0A1Y1MNM7 A0A1Q3FX89 A0A151WQ01 F4X5V0 A0A084W692 A0A154PLC9 A0A067RPE4 A0A2M4AQ35 A0A2M4AEX4 A0A034VJS4 A0A2M4AEQ0 A0A2A3EK57 A0A0A1X4X1 A0A2M4ADW3 A0A1L8E385 A0A0K8U1Q9 A0A0K8TMV1 W8CB69 A0A2M4CV67 A0A2M4CUX5 A0A1B0G2Z2 E9IEG0 A0A151JM01 A0A1A9X1P2 A0A336LSQ6 A0A336MNJ6 A0A151JVK9 A0A1B6CFN2 A0A0L7RCA1 A0A195CDY2 A0A1I8NTD7 A0A1I8NTD9 E2B7S6 A0A182QZ66 Q7QII1 A0A1B6MKD5 A0A1I8ME42 A0A1I8ME39 K7IXZ4 A0A1B6IDQ5 A0A336LN84 A0A146M6N0 A0A1B6K5F8 A0A1A9UQW6 B4NGP5 A0A0A9XLD6 A0A0T6B7K0 E2A3U5 A0A1B0BTE7 A0A069DU90 A0A182Y833 A0A182PIP7 A0A182RJE7 A0A182U2D9 A0A182I6K0 A0A182L3V4 A0A182VMB5 A0A182K2D1 A0A2M4BJ10 A0A1A9XK02 A0A1A9ZWR5 A0A023F450 R7U746 A0A1B0CCI3 A0A182FZP5 A0A1S3K926 B4HHK5 B4K5I6 V5H9U6 A0A182J7T1 B4JIK8 A0A1C9TA51 B4LWE0 A0A0B4KGZ8 B3LYA8 Q9VC35 B4QTS6
Pubmed
22118469
26354079
23622113
24508170
30249741
18362917
+ More
19820115 17510324 26999592 24945155 28004739 21719571 24438588 24845553 25348373 25830018 26369729 24495485 21282665 20798317 12364791 14747013 17210077 25315136 20075255 26823975 17994087 25401762 26334808 25244985 20966253 25474469 23254933 25765539 18574105 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10731138 26512105
19820115 17510324 26999592 24945155 28004739 21719571 24438588 24845553 25348373 25830018 26369729 24495485 21282665 20798317 12364791 14747013 17210077 25315136 20075255 26823975 17994087 25401762 26334808 25244985 20966253 25474469 23254933 25765539 18574105 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10731138 26512105
EMBL
RSAL01000222
RVE44020.1
AGBW02010898
OWR47558.1
GDQN01003491
JAT87563.1
+ More
KQ459984 KPJ19018.1 KQ459220 KPJ02899.1 GAIX01004953 JAA87607.1 GEHC01000120 JAV47525.1 ODYU01009018 SOQ53111.1 NEVH01006570 PNF37816.1 GALX01004445 JAB64021.1 GALX01004444 JAB64022.1 KK107238 QOIP01000007 EZA54789.1 RLU20537.1 GFDL01001098 JAV33947.1 KQ971377 KYB25110.1 CH477627 EAT38110.1 EEZ97380.2 GFDL01002890 JAV32155.1 GBYB01002241 JAG72008.1 GDUN01000544 JAN95375.1 GAPW01000636 JAC12962.1 GFDL01001101 JAV33944.1 GEZM01026059 GEZM01026058 JAV87269.1 GFDL01002887 JAV32158.1 KQ982865 KYQ49765.1 GL888742 EGI58190.1 ATLV01020760 ATLV01020761 ATLV01020762 KE525307 KFB45736.1 KQ434960 KZC12675.1 KK852505 KDR22505.1 GGFK01009585 MBW42906.1 GGFK01006015 MBW39336.1 GAKP01015416 GAKP01015403 GAKP01015401 JAC43551.1 GGFK01005933 MBW39254.1 KZ288226 PBC31874.1 GBXI01007938 JAD06354.1 GGFK01005648 MBW38969.1 GFDF01001042 JAV13042.1 GDHF01031630 JAI20684.1 GDAI01002132 JAI15471.1 GAMC01005111 JAC01445.1 GGFL01004977 MBW69155.1 GGFL01004976 MBW69154.1 CCAG010003320 GL762591 EFZ21031.1 KQ979011 KYN26637.1 UFQT01000086 SSX19457.1 UFQT01001813 SSX31876.1 KQ981706 KYN37420.1 GEDC01025077 JAS12221.1 KQ414616 KOC68425.1 KQ977957 KYM98446.1 GL446209 EFN88219.1 AXCN02000560 AAAB01008807 EAA04252.3 GEBQ01003540 JAT36437.1 GECU01022652 JAS85054.1 SSX19456.1 GDHC01012665 GDHC01004223 JAQ05964.1 JAQ14406.1 GECU01001010 JAT06697.1 CH964272 EDW84392.1 GBHO01025664 GBHO01025663 JAG17940.1 JAG17941.1 LJIG01009317 KRT83322.1 GL436500 EFN71889.1 JXJN01020099 JXJN01020100 GBGD01001201 JAC87688.1 APCN01003655 GGFJ01003657 MBW52798.1 GBBI01002943 JAC15769.1 AMQN01010248 KB307403 ELT98960.1 AJWK01006813 AJWK01006814 CH480815 EDW43547.1 CH933806 EDW14023.1 GANP01010534 JAB73934.1 CH916369 EDV93089.1 KU553315 AOR07013.1 CH940650 EDW66577.2 AE014297 AGB96310.1 CH902617 EDV44012.1 AF132157 BT003212 BT057989 AAD34745.1 AAO24967.1 ACM16699.1 CM000364 EDX14280.1
KQ459984 KPJ19018.1 KQ459220 KPJ02899.1 GAIX01004953 JAA87607.1 GEHC01000120 JAV47525.1 ODYU01009018 SOQ53111.1 NEVH01006570 PNF37816.1 GALX01004445 JAB64021.1 GALX01004444 JAB64022.1 KK107238 QOIP01000007 EZA54789.1 RLU20537.1 GFDL01001098 JAV33947.1 KQ971377 KYB25110.1 CH477627 EAT38110.1 EEZ97380.2 GFDL01002890 JAV32155.1 GBYB01002241 JAG72008.1 GDUN01000544 JAN95375.1 GAPW01000636 JAC12962.1 GFDL01001101 JAV33944.1 GEZM01026059 GEZM01026058 JAV87269.1 GFDL01002887 JAV32158.1 KQ982865 KYQ49765.1 GL888742 EGI58190.1 ATLV01020760 ATLV01020761 ATLV01020762 KE525307 KFB45736.1 KQ434960 KZC12675.1 KK852505 KDR22505.1 GGFK01009585 MBW42906.1 GGFK01006015 MBW39336.1 GAKP01015416 GAKP01015403 GAKP01015401 JAC43551.1 GGFK01005933 MBW39254.1 KZ288226 PBC31874.1 GBXI01007938 JAD06354.1 GGFK01005648 MBW38969.1 GFDF01001042 JAV13042.1 GDHF01031630 JAI20684.1 GDAI01002132 JAI15471.1 GAMC01005111 JAC01445.1 GGFL01004977 MBW69155.1 GGFL01004976 MBW69154.1 CCAG010003320 GL762591 EFZ21031.1 KQ979011 KYN26637.1 UFQT01000086 SSX19457.1 UFQT01001813 SSX31876.1 KQ981706 KYN37420.1 GEDC01025077 JAS12221.1 KQ414616 KOC68425.1 KQ977957 KYM98446.1 GL446209 EFN88219.1 AXCN02000560 AAAB01008807 EAA04252.3 GEBQ01003540 JAT36437.1 GECU01022652 JAS85054.1 SSX19456.1 GDHC01012665 GDHC01004223 JAQ05964.1 JAQ14406.1 GECU01001010 JAT06697.1 CH964272 EDW84392.1 GBHO01025664 GBHO01025663 JAG17940.1 JAG17941.1 LJIG01009317 KRT83322.1 GL436500 EFN71889.1 JXJN01020099 JXJN01020100 GBGD01001201 JAC87688.1 APCN01003655 GGFJ01003657 MBW52798.1 GBBI01002943 JAC15769.1 AMQN01010248 KB307403 ELT98960.1 AJWK01006813 AJWK01006814 CH480815 EDW43547.1 CH933806 EDW14023.1 GANP01010534 JAB73934.1 CH916369 EDV93089.1 KU553315 AOR07013.1 CH940650 EDW66577.2 AE014297 AGB96310.1 CH902617 EDV44012.1 AF132157 BT003212 BT057989 AAD34745.1 AAO24967.1 ACM16699.1 CM000364 EDX14280.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000053268
UP000235965
UP000005203
+ More
UP000053097 UP000279307 UP000007266 UP000008820 UP000075809 UP000007755 UP000030765 UP000076502 UP000027135 UP000242457 UP000092444 UP000078492 UP000091820 UP000078541 UP000053825 UP000078542 UP000095300 UP000008237 UP000075886 UP000007062 UP000095301 UP000002358 UP000078200 UP000007798 UP000000311 UP000092460 UP000076408 UP000075885 UP000075900 UP000075902 UP000075840 UP000075882 UP000075903 UP000075881 UP000092443 UP000092445 UP000014760 UP000092461 UP000069272 UP000085678 UP000001292 UP000009192 UP000075880 UP000001070 UP000008792 UP000000803 UP000007801 UP000000304
UP000053097 UP000279307 UP000007266 UP000008820 UP000075809 UP000007755 UP000030765 UP000076502 UP000027135 UP000242457 UP000092444 UP000078492 UP000091820 UP000078541 UP000053825 UP000078542 UP000095300 UP000008237 UP000075886 UP000007062 UP000095301 UP000002358 UP000078200 UP000007798 UP000000311 UP000092460 UP000076408 UP000075885 UP000075900 UP000075902 UP000075840 UP000075882 UP000075903 UP000075881 UP000092443 UP000092445 UP000014760 UP000092461 UP000069272 UP000085678 UP000001292 UP000009192 UP000075880 UP000001070 UP000008792 UP000000803 UP000007801 UP000000304
PRIDE
Pfam
PF04791 LMBR1
ProteinModelPortal
A0A3S2NN29
A0A212F1E0
A0A1E1WKM5
A0A194RMG5
A0A194QBV4
S4PXJ0
+ More
A0A1W7R8U3 A0A2H1WJ69 A0A2J7RAF3 A0A087ZQT2 V5I8M1 V5GR09 A0A026WI55 A0A1Q3G2A0 A0A139WB21 Q16UA7 D6X3S1 A0A1Q3FX73 A0A0C9R641 A0A0P6K0Y0 A0A023EVE3 A0A1Q3G2B7 A0A1Y1MNM7 A0A1Q3FX89 A0A151WQ01 F4X5V0 A0A084W692 A0A154PLC9 A0A067RPE4 A0A2M4AQ35 A0A2M4AEX4 A0A034VJS4 A0A2M4AEQ0 A0A2A3EK57 A0A0A1X4X1 A0A2M4ADW3 A0A1L8E385 A0A0K8U1Q9 A0A0K8TMV1 W8CB69 A0A2M4CV67 A0A2M4CUX5 A0A1B0G2Z2 E9IEG0 A0A151JM01 A0A1A9X1P2 A0A336LSQ6 A0A336MNJ6 A0A151JVK9 A0A1B6CFN2 A0A0L7RCA1 A0A195CDY2 A0A1I8NTD7 A0A1I8NTD9 E2B7S6 A0A182QZ66 Q7QII1 A0A1B6MKD5 A0A1I8ME42 A0A1I8ME39 K7IXZ4 A0A1B6IDQ5 A0A336LN84 A0A146M6N0 A0A1B6K5F8 A0A1A9UQW6 B4NGP5 A0A0A9XLD6 A0A0T6B7K0 E2A3U5 A0A1B0BTE7 A0A069DU90 A0A182Y833 A0A182PIP7 A0A182RJE7 A0A182U2D9 A0A182I6K0 A0A182L3V4 A0A182VMB5 A0A182K2D1 A0A2M4BJ10 A0A1A9XK02 A0A1A9ZWR5 A0A023F450 R7U746 A0A1B0CCI3 A0A182FZP5 A0A1S3K926 B4HHK5 B4K5I6 V5H9U6 A0A182J7T1 B4JIK8 A0A1C9TA51 B4LWE0 A0A0B4KGZ8 B3LYA8 Q9VC35 B4QTS6
A0A1W7R8U3 A0A2H1WJ69 A0A2J7RAF3 A0A087ZQT2 V5I8M1 V5GR09 A0A026WI55 A0A1Q3G2A0 A0A139WB21 Q16UA7 D6X3S1 A0A1Q3FX73 A0A0C9R641 A0A0P6K0Y0 A0A023EVE3 A0A1Q3G2B7 A0A1Y1MNM7 A0A1Q3FX89 A0A151WQ01 F4X5V0 A0A084W692 A0A154PLC9 A0A067RPE4 A0A2M4AQ35 A0A2M4AEX4 A0A034VJS4 A0A2M4AEQ0 A0A2A3EK57 A0A0A1X4X1 A0A2M4ADW3 A0A1L8E385 A0A0K8U1Q9 A0A0K8TMV1 W8CB69 A0A2M4CV67 A0A2M4CUX5 A0A1B0G2Z2 E9IEG0 A0A151JM01 A0A1A9X1P2 A0A336LSQ6 A0A336MNJ6 A0A151JVK9 A0A1B6CFN2 A0A0L7RCA1 A0A195CDY2 A0A1I8NTD7 A0A1I8NTD9 E2B7S6 A0A182QZ66 Q7QII1 A0A1B6MKD5 A0A1I8ME42 A0A1I8ME39 K7IXZ4 A0A1B6IDQ5 A0A336LN84 A0A146M6N0 A0A1B6K5F8 A0A1A9UQW6 B4NGP5 A0A0A9XLD6 A0A0T6B7K0 E2A3U5 A0A1B0BTE7 A0A069DU90 A0A182Y833 A0A182PIP7 A0A182RJE7 A0A182U2D9 A0A182I6K0 A0A182L3V4 A0A182VMB5 A0A182K2D1 A0A2M4BJ10 A0A1A9XK02 A0A1A9ZWR5 A0A023F450 R7U746 A0A1B0CCI3 A0A182FZP5 A0A1S3K926 B4HHK5 B4K5I6 V5H9U6 A0A182J7T1 B4JIK8 A0A1C9TA51 B4LWE0 A0A0B4KGZ8 B3LYA8 Q9VC35 B4QTS6
Ontologies
PANTHER
Topology
Subcellular location
Cell membrane
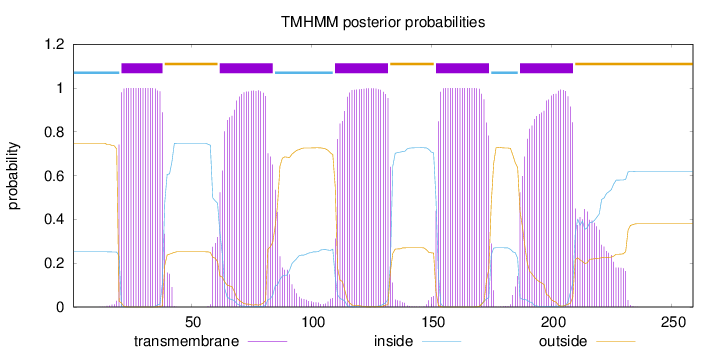
Length:
259
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
114.91316
Exp number, first 60 AAs:
19.96132
Total prob of N-in:
0.25222
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 38
outside
39 - 61
TMhelix
62 - 84
inside
85 - 109
TMhelix
110 - 132
outside
133 - 151
TMhelix
152 - 174
inside
175 - 186
TMhelix
187 - 209
outside
210 - 259
Population Genetic Test Statistics
Pi
186.473454
Theta
167.505449
Tajima's D
0.212723
CLR
0.004273
CSRT
0.426228688565572
Interpretation
Uncertain
