Pre Gene Modal
BGIBMGA002416
Annotation
PREDICTED:_uncharacterized_protein_LOC101746538_[Bombyx_mori]
Full name
MICOS complex subunit MIC13
+ More
MICOS complex subunit MIC13 homolog QIL1
MICOS complex subunit MIC13 homolog QIL1
Location in the cell
Extracellular Reliability : 1.306 Mitochondrial Reliability : 1.072
Sequence
CDS
ATGTTAAAGTTTGGCGTGAAATCTGTGTTACTTGGATCCGCAGTGTACTACACAATAGATAAAGGTGTGTGGAAAGATAGTGCTACGACAGCGGCGATATATGATGAACTTGAAAAAGGAATGTCACCATATGTCGGGGAACTAAAGAGCCAAGTTCCATACGAACTACCAGCATTACCTTCTAATGACAGGATATCATATTTGTTCAAGTACTACTGGAATTGCGGTGTTAAAGCTACATTTAGATTTTTAGTCGAATTGCCTACCCATACTAGTAATGCAGCTTTCAAGACATACGACTTTATTTCATCATCTTTGGAAACTCCAGAACCACATCAAGAAAAAGATAAATGA
Protein
MLKFGVKSVLLGSAVYYTIDKGVWKDSATTAAIYDELEKGMSPYVGELKSQVPYELPALPSNDRISYLFKYYWNCGVKATFRFLVELPTHTSNAAFKTYDFISSSLETPEPHQEKDK
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Similarity
Belongs to the MICOS complex subunit Mic13 family.
Keywords
Complete proteome
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain MICOS complex subunit MIC13 homolog QIL1
Uniprot
A0A1E1VZP0
S4NSC0
A0A212F1H4
A0A0L7LMX3
A0A3S2N837
A0A194RNM5
+ More
I4DKS9 A0A0T6B8U9 A0A1B0DMH3 N6UEB3 U4U4E9 A0A3Q0JDD6 A0A023EDK2 D7EIA3 A0A067RRJ5 A0A1B0CH57 A0A023EFH1 A0A182KAE9 A0A1B2AJ92 Q9VVH3 A0A182JEA2 T1D5D1 B3NHV0 E0VB41 A0A1Y1LTK4 W5JVB3 A0A1Q3FK34 A0A182YFL7 B4QNV3 T1E8I2 A0A2M3Z730 A0A182XJ81 A0A182LBC0 Q7Q9Q1 B4HKS8 A0A182HP83 A0A1L8DC16 B4PJV1 A0A182PHC3 W8BGK8 B4N6F6 Q1HRM0 B0X7Y1 A0A182WA96 A0A2M4C3M3 A0A2M4C3Q7 A0A2M4C3P1 A0A2M4C3M2 A0A182M717 B0XB56 Q2LZI2 A0A182FCF5 A0A3B0KQ46 A0A182N821 A0A084W2J7 A0A2A3ER28 B4KVC3 A0A0P4VLZ0 A0A034WQA4 A0A2M4AJ48 B4H6M8 A0A2R7W6X5 A0A0L0CGZ5 A0A1A9WAA0 A0A158NLG6 A0A182RTM9 A0A088AG05 A0A2M4AJC8 A0A026WYD3 A0A0K8TYZ5 A0A1B0A2N7 A0A1W4VRT9 A0A0A9ZGK4 A0A158NLG5 A0A195B2E1 B4LBC6 A0A1I8M4F5 A0A023FA41 A0A1L8EEJ6 A0A195E6C3 B4IXT4 B3M840 U5ETV4 A0A1L8EEI2 A0A1I8NYI6 F4WVY0 A0A195F2V4 A0A069DMN6 A0A0V0G9K8 A0A151XCK0 A0A224Y0Z0 R4WJQ6 E9IU47 A0A195CYB9 K7J128 A0A0M4EMU4 A0A1B6CSR8 A0A232FMV9 A0A336KSX5
I4DKS9 A0A0T6B8U9 A0A1B0DMH3 N6UEB3 U4U4E9 A0A3Q0JDD6 A0A023EDK2 D7EIA3 A0A067RRJ5 A0A1B0CH57 A0A023EFH1 A0A182KAE9 A0A1B2AJ92 Q9VVH3 A0A182JEA2 T1D5D1 B3NHV0 E0VB41 A0A1Y1LTK4 W5JVB3 A0A1Q3FK34 A0A182YFL7 B4QNV3 T1E8I2 A0A2M3Z730 A0A182XJ81 A0A182LBC0 Q7Q9Q1 B4HKS8 A0A182HP83 A0A1L8DC16 B4PJV1 A0A182PHC3 W8BGK8 B4N6F6 Q1HRM0 B0X7Y1 A0A182WA96 A0A2M4C3M3 A0A2M4C3Q7 A0A2M4C3P1 A0A2M4C3M2 A0A182M717 B0XB56 Q2LZI2 A0A182FCF5 A0A3B0KQ46 A0A182N821 A0A084W2J7 A0A2A3ER28 B4KVC3 A0A0P4VLZ0 A0A034WQA4 A0A2M4AJ48 B4H6M8 A0A2R7W6X5 A0A0L0CGZ5 A0A1A9WAA0 A0A158NLG6 A0A182RTM9 A0A088AG05 A0A2M4AJC8 A0A026WYD3 A0A0K8TYZ5 A0A1B0A2N7 A0A1W4VRT9 A0A0A9ZGK4 A0A158NLG5 A0A195B2E1 B4LBC6 A0A1I8M4F5 A0A023FA41 A0A1L8EEJ6 A0A195E6C3 B4IXT4 B3M840 U5ETV4 A0A1L8EEI2 A0A1I8NYI6 F4WVY0 A0A195F2V4 A0A069DMN6 A0A0V0G9K8 A0A151XCK0 A0A224Y0Z0 R4WJQ6 E9IU47 A0A195CYB9 K7J128 A0A0M4EMU4 A0A1B6CSR8 A0A232FMV9 A0A336KSX5
Pubmed
23622113
22118469
26227816
26354079
22651552
23537049
+ More
24945155 18362917 19820115 24845553 26483478 10731132 12537572 12537569 25997101 24330624 17994087 20566863 28004739 20920257 23761445 25244985 22936249 20966253 12364791 14747013 17210077 17550304 24495485 17204158 17510324 15632085 24438588 27129103 25348373 26108605 21347285 24508170 30249741 25401762 26823975 25315136 25474469 21719571 26334808 23691247 21282665 20075255 28648823
24945155 18362917 19820115 24845553 26483478 10731132 12537572 12537569 25997101 24330624 17994087 20566863 28004739 20920257 23761445 25244985 22936249 20966253 12364791 14747013 17210077 17550304 24495485 17204158 17510324 15632085 24438588 27129103 25348373 26108605 21347285 24508170 30249741 25401762 26823975 25315136 25474469 21719571 26334808 23691247 21282665 20075255 28648823
EMBL
GDQN01010854
JAT80200.1
GAIX01014162
JAA78398.1
AGBW02010898
OWR47563.1
+ More
JTDY01000499 KOB76903.1 RSAL01000222 RVE44017.1 KQ459984 KPJ19014.1 AK401897 KQ459220 BAM18519.1 KPJ02896.1 LJIG01009098 KRT83760.1 AJVK01001342 APGK01038500 KB740960 ENN76992.1 KB631698 ERL85466.1 GAPW01006055 JAC07543.1 DS497675 EFA11746.1 KK852470 KDR23245.1 AJWK01002882 AJWK01012065 JXUM01077074 JXUM01080018 GAPW01006058 GAPW01006057 GAPW01006056 KQ563159 KQ562985 JAC07540.1 KXJ74377.1 KXJ74706.1 KX531412 ANY27222.1 AE014296 AY071567 GALA01000573 JAA94279.1 CH954178 EDV51965.1 DS235021 EEB10597.1 GEZM01046870 JAV76982.1 ADMH02000391 ETN66739.1 GFDL01007160 JAV27885.1 CM000363 CM002912 EDX10855.1 KMZ00235.1 GAMD01002743 JAA98847.1 GGFM01003497 MBW24248.1 AAAB01008900 EAA09512.4 CH480815 EDW41883.1 APCN01002923 GFDF01010076 JAV04008.1 CM000159 EDW94720.1 GAMC01017763 GAMC01017761 JAB88794.1 CH964154 EDW79945.1 DQ440074 CH477862 ABF18107.1 EAT35522.1 DS232468 EDS42200.1 GGFJ01010761 MBW59902.1 GGFJ01010764 MBW59905.1 GGFJ01010763 MBW59904.1 GGFJ01010762 MBW59903.1 AXCM01006295 DS232614 EDS44109.1 CH379069 EAL29525.1 OUUW01000012 SPP87321.1 ATLV01019621 KE525275 KFB44441.1 KZ288197 PBC33722.1 CH933809 EDW18366.1 GDKW01003137 JAI53458.1 GAKP01003014 JAC55938.1 GGFK01007483 MBW40804.1 CH479214 EDW33456.1 KK854267 PTY13965.1 JRES01000413 KNC31467.1 ADTU01019399 GGFK01007565 MBW40886.1 KK107063 QOIP01000005 EZA61047.1 RLU22809.1 GDHF01032615 JAI19699.1 GBHO01002499 GBRD01000961 GDHC01008397 GDHC01002382 JAG41105.1 JAG64860.1 JAQ10232.1 JAQ16247.1 KQ976662 KYM78369.1 CH940647 EDW69714.1 GBBI01000442 JAC18270.1 GFDG01001679 JAV17120.1 KQ979568 KYN20723.1 CH916366 EDV96455.1 CH902618 EDV39948.1 GANO01002611 JAB57260.1 GFDG01001677 JAV17122.1 GL888404 EGI61569.1 KQ981864 KYN34419.1 GBGD01003566 JAC85323.1 GECL01002077 JAP04047.1 KQ982314 KYQ58069.1 GFTR01001756 JAW14670.1 AK417815 BAN21030.1 GL765841 EFZ15926.1 KQ977115 KYN05660.1 CP012525 ALC43130.1 GEDC01020709 JAS16589.1 NNAY01000009 OXU32071.1 UFQS01000742 UFQT01000742 SSX06515.1 SSX26864.1
JTDY01000499 KOB76903.1 RSAL01000222 RVE44017.1 KQ459984 KPJ19014.1 AK401897 KQ459220 BAM18519.1 KPJ02896.1 LJIG01009098 KRT83760.1 AJVK01001342 APGK01038500 KB740960 ENN76992.1 KB631698 ERL85466.1 GAPW01006055 JAC07543.1 DS497675 EFA11746.1 KK852470 KDR23245.1 AJWK01002882 AJWK01012065 JXUM01077074 JXUM01080018 GAPW01006058 GAPW01006057 GAPW01006056 KQ563159 KQ562985 JAC07540.1 KXJ74377.1 KXJ74706.1 KX531412 ANY27222.1 AE014296 AY071567 GALA01000573 JAA94279.1 CH954178 EDV51965.1 DS235021 EEB10597.1 GEZM01046870 JAV76982.1 ADMH02000391 ETN66739.1 GFDL01007160 JAV27885.1 CM000363 CM002912 EDX10855.1 KMZ00235.1 GAMD01002743 JAA98847.1 GGFM01003497 MBW24248.1 AAAB01008900 EAA09512.4 CH480815 EDW41883.1 APCN01002923 GFDF01010076 JAV04008.1 CM000159 EDW94720.1 GAMC01017763 GAMC01017761 JAB88794.1 CH964154 EDW79945.1 DQ440074 CH477862 ABF18107.1 EAT35522.1 DS232468 EDS42200.1 GGFJ01010761 MBW59902.1 GGFJ01010764 MBW59905.1 GGFJ01010763 MBW59904.1 GGFJ01010762 MBW59903.1 AXCM01006295 DS232614 EDS44109.1 CH379069 EAL29525.1 OUUW01000012 SPP87321.1 ATLV01019621 KE525275 KFB44441.1 KZ288197 PBC33722.1 CH933809 EDW18366.1 GDKW01003137 JAI53458.1 GAKP01003014 JAC55938.1 GGFK01007483 MBW40804.1 CH479214 EDW33456.1 KK854267 PTY13965.1 JRES01000413 KNC31467.1 ADTU01019399 GGFK01007565 MBW40886.1 KK107063 QOIP01000005 EZA61047.1 RLU22809.1 GDHF01032615 JAI19699.1 GBHO01002499 GBRD01000961 GDHC01008397 GDHC01002382 JAG41105.1 JAG64860.1 JAQ10232.1 JAQ16247.1 KQ976662 KYM78369.1 CH940647 EDW69714.1 GBBI01000442 JAC18270.1 GFDG01001679 JAV17120.1 KQ979568 KYN20723.1 CH916366 EDV96455.1 CH902618 EDV39948.1 GANO01002611 JAB57260.1 GFDG01001677 JAV17122.1 GL888404 EGI61569.1 KQ981864 KYN34419.1 GBGD01003566 JAC85323.1 GECL01002077 JAP04047.1 KQ982314 KYQ58069.1 GFTR01001756 JAW14670.1 AK417815 BAN21030.1 GL765841 EFZ15926.1 KQ977115 KYN05660.1 CP012525 ALC43130.1 GEDC01020709 JAS16589.1 NNAY01000009 OXU32071.1 UFQS01000742 UFQT01000742 SSX06515.1 SSX26864.1
Proteomes
UP000007151
UP000037510
UP000283053
UP000053240
UP000053268
UP000092462
+ More
UP000019118 UP000030742 UP000079169 UP000007266 UP000027135 UP000092461 UP000069940 UP000249989 UP000075881 UP000000803 UP000075880 UP000008711 UP000009046 UP000000673 UP000076408 UP000000304 UP000076407 UP000075882 UP000007062 UP000001292 UP000075840 UP000002282 UP000075885 UP000007798 UP000008820 UP000002320 UP000075920 UP000075883 UP000001819 UP000069272 UP000268350 UP000075884 UP000030765 UP000242457 UP000009192 UP000008744 UP000037069 UP000091820 UP000005205 UP000075900 UP000005203 UP000053097 UP000279307 UP000092445 UP000192221 UP000078540 UP000008792 UP000095301 UP000078492 UP000001070 UP000007801 UP000095300 UP000007755 UP000078541 UP000075809 UP000078542 UP000002358 UP000092553 UP000215335
UP000019118 UP000030742 UP000079169 UP000007266 UP000027135 UP000092461 UP000069940 UP000249989 UP000075881 UP000000803 UP000075880 UP000008711 UP000009046 UP000000673 UP000076408 UP000000304 UP000076407 UP000075882 UP000007062 UP000001292 UP000075840 UP000002282 UP000075885 UP000007798 UP000008820 UP000002320 UP000075920 UP000075883 UP000001819 UP000069272 UP000268350 UP000075884 UP000030765 UP000242457 UP000009192 UP000008744 UP000037069 UP000091820 UP000005205 UP000075900 UP000005203 UP000053097 UP000279307 UP000092445 UP000192221 UP000078540 UP000008792 UP000095301 UP000078492 UP000001070 UP000007801 UP000095300 UP000007755 UP000078541 UP000075809 UP000078542 UP000002358 UP000092553 UP000215335
PRIDE
Pfam
PF15884 QIL1
Interpro
IPR026769
Mic13
ProteinModelPortal
A0A1E1VZP0
S4NSC0
A0A212F1H4
A0A0L7LMX3
A0A3S2N837
A0A194RNM5
+ More
I4DKS9 A0A0T6B8U9 A0A1B0DMH3 N6UEB3 U4U4E9 A0A3Q0JDD6 A0A023EDK2 D7EIA3 A0A067RRJ5 A0A1B0CH57 A0A023EFH1 A0A182KAE9 A0A1B2AJ92 Q9VVH3 A0A182JEA2 T1D5D1 B3NHV0 E0VB41 A0A1Y1LTK4 W5JVB3 A0A1Q3FK34 A0A182YFL7 B4QNV3 T1E8I2 A0A2M3Z730 A0A182XJ81 A0A182LBC0 Q7Q9Q1 B4HKS8 A0A182HP83 A0A1L8DC16 B4PJV1 A0A182PHC3 W8BGK8 B4N6F6 Q1HRM0 B0X7Y1 A0A182WA96 A0A2M4C3M3 A0A2M4C3Q7 A0A2M4C3P1 A0A2M4C3M2 A0A182M717 B0XB56 Q2LZI2 A0A182FCF5 A0A3B0KQ46 A0A182N821 A0A084W2J7 A0A2A3ER28 B4KVC3 A0A0P4VLZ0 A0A034WQA4 A0A2M4AJ48 B4H6M8 A0A2R7W6X5 A0A0L0CGZ5 A0A1A9WAA0 A0A158NLG6 A0A182RTM9 A0A088AG05 A0A2M4AJC8 A0A026WYD3 A0A0K8TYZ5 A0A1B0A2N7 A0A1W4VRT9 A0A0A9ZGK4 A0A158NLG5 A0A195B2E1 B4LBC6 A0A1I8M4F5 A0A023FA41 A0A1L8EEJ6 A0A195E6C3 B4IXT4 B3M840 U5ETV4 A0A1L8EEI2 A0A1I8NYI6 F4WVY0 A0A195F2V4 A0A069DMN6 A0A0V0G9K8 A0A151XCK0 A0A224Y0Z0 R4WJQ6 E9IU47 A0A195CYB9 K7J128 A0A0M4EMU4 A0A1B6CSR8 A0A232FMV9 A0A336KSX5
I4DKS9 A0A0T6B8U9 A0A1B0DMH3 N6UEB3 U4U4E9 A0A3Q0JDD6 A0A023EDK2 D7EIA3 A0A067RRJ5 A0A1B0CH57 A0A023EFH1 A0A182KAE9 A0A1B2AJ92 Q9VVH3 A0A182JEA2 T1D5D1 B3NHV0 E0VB41 A0A1Y1LTK4 W5JVB3 A0A1Q3FK34 A0A182YFL7 B4QNV3 T1E8I2 A0A2M3Z730 A0A182XJ81 A0A182LBC0 Q7Q9Q1 B4HKS8 A0A182HP83 A0A1L8DC16 B4PJV1 A0A182PHC3 W8BGK8 B4N6F6 Q1HRM0 B0X7Y1 A0A182WA96 A0A2M4C3M3 A0A2M4C3Q7 A0A2M4C3P1 A0A2M4C3M2 A0A182M717 B0XB56 Q2LZI2 A0A182FCF5 A0A3B0KQ46 A0A182N821 A0A084W2J7 A0A2A3ER28 B4KVC3 A0A0P4VLZ0 A0A034WQA4 A0A2M4AJ48 B4H6M8 A0A2R7W6X5 A0A0L0CGZ5 A0A1A9WAA0 A0A158NLG6 A0A182RTM9 A0A088AG05 A0A2M4AJC8 A0A026WYD3 A0A0K8TYZ5 A0A1B0A2N7 A0A1W4VRT9 A0A0A9ZGK4 A0A158NLG5 A0A195B2E1 B4LBC6 A0A1I8M4F5 A0A023FA41 A0A1L8EEJ6 A0A195E6C3 B4IXT4 B3M840 U5ETV4 A0A1L8EEI2 A0A1I8NYI6 F4WVY0 A0A195F2V4 A0A069DMN6 A0A0V0G9K8 A0A151XCK0 A0A224Y0Z0 R4WJQ6 E9IU47 A0A195CYB9 K7J128 A0A0M4EMU4 A0A1B6CSR8 A0A232FMV9 A0A336KSX5
Ontologies
PANTHER
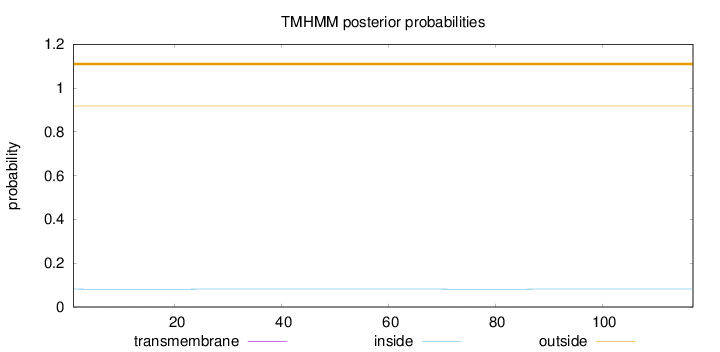
Topology
Subcellular location
Mitochondrion inner membrane
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0145
Exp number, first 60 AAs:
0.00895
Total prob of N-in:
0.08174
outside
1 - 117
Population Genetic Test Statistics
Pi
239.857007
Theta
198.901117
Tajima's D
0.659024
CLR
44.982659
CSRT
0.56277186140693
Interpretation
Uncertain
